Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants
- PMID: 34016740
- PMCID: PMC8284396
- DOI: 10.1126/science.abh1139
Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants
Abstract
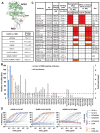
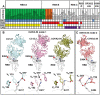
Neutralizing antibodies (nAbs) elicited against the receptor binding site (RBS) of the spike protein of wild-type severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are generally less effective against recent variants of concern. RBS residues Glu484, Lys417, and Asn501 are mutated in variants first described in South Africa (B.1.351) and Brazil (P.1). We analyzed their effects on angiotensin-converting enzyme 2 binding, as well as the effects of two of these mutations (K417N and E484K) on nAbs isolated from COVID-19 patients. Binding and neutralization of the two most frequently elicited antibody families (IGHV3-53/3-66 and IGHV1-2), which can both bind the RBS in alternative binding modes, are abrogated by K417N, E484K, or both. These effects can be structurally explained by their extensive interactions with RBS nAbs. However, nAbs to the more conserved, cross-neutralizing CR3022 and S309 sites were largely unaffected. The results have implications for next-generation vaccines and antibody therapies.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

Update of
-
Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants.bioRxiv [Preprint]. 2021 Feb 17:2021.02.16.430500. doi: 10.1101/2021.02.16.430500. bioRxiv. 2021. Update in: Science. 2021 Aug 13;373(6556):818-823. doi: 10.1126/science.abh1139 PMID: 33619487 Free PMC article. Updated. Preprint.
Similar articles
-
Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants.bioRxiv [Preprint]. 2021 Feb 17:2021.02.16.430500. doi: 10.1101/2021.02.16.430500. bioRxiv. 2021. Update in: Science. 2021 Aug 13;373(6556):818-823. doi: 10.1126/science.abh1139 PMID: 33619487 Free PMC article. Updated. Preprint.
-
Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies.Nature. 2021 May;593(7857):136-141. doi: 10.1038/s41586-021-03412-7. Epub 2021 Mar 11. Nature. 2021. PMID: 33706364 Free PMC article.
-
Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.Science. 2021 Feb 12;371(6530):eabe6230. doi: 10.1126/science.abe6230. Epub 2021 Jan 12. Science. 2021. PMID: 33436526 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
50 Years of structural immunology.J Biol Chem. 2021 Jan-Jun;296:100745. doi: 10.1016/j.jbc.2021.100745. Epub 2021 May 3. J Biol Chem. 2021. PMID: 33957119 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.Nat Microbiol. 2022 Oct;7(10):1635-1649. doi: 10.1038/s41564-022-01235-4. Epub 2022 Sep 23. Nat Microbiol. 2022. PMID: 36151403 Free PMC article.
-
The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape.medRxiv [Preprint]. 2021 Jul 25:2021.02.23.21252268. doi: 10.1101/2021.02.23.21252268. medRxiv. 2021. Update in: Cell. 2021 Sep 30;184(20):5189-5200.e7. doi: 10.1016/j.cell.2021.09.003 PMID: 33688681 Free PMC article. Updated. Preprint.
-
Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection.Cell Rep Med. 2022 Jan 24;3(2):100528. doi: 10.1016/j.xcrm.2022.100528. eCollection 2022 Feb 15. Cell Rep Med. 2022. PMID: 35233549 Free PMC article.
-
A CNN model for predicting binding affinity changes between SARS-CoV-2 spike RBD variants and ACE2 homologues.bioRxiv [Preprint]. 2022 Mar 23:2022.03.22.485413. doi: 10.1101/2022.03.22.485413. bioRxiv. 2022. PMID: 35350198 Free PMC article. Preprint.
-
A Pre-Vaccination Baseline of SARS-CoV-2 Genetic Surveillance and Diversity in the United States.Viruses. 2022 Jan 7;14(1):104. doi: 10.3390/v14010104. Viruses. 2022. PMID: 35062308 Free PMC article.
References
-
- M. Chand et al., Investigation of Novel SARS-CoV-2 Variant: Variant of Concern 202012/01. Technical Briefing 5. Public Health England (2020); https://assets.publishing.service.gov.uk/government/uploads/system/uploa....
-
- H. Tegally et al., Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv [preprint]. 22 December 2020. - PubMed
-
- N. R. Faria et al., “Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings” (2021); https://virological.org/t/genomic-characterisation-of-an-emergent-sars-c....
-
- V. Tchesnokova, H. Kulakesara, L. Larson, V. Bowers, E. Rechkina, D. Kisiela, Y. Sledneva, D. Choudhury, I. Maslova, K. Deng, K. Kutumbaka, H. Geng, C. Fowler, D. Greene, J. Ralston, M. Samadpour, E. Sokurenko, Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-Cov-2 variants. bioRxiv [preprint]. 22 February 2021. - PMC - PubMed
-
- Yadav P. D., Sapkal G. N., Abraham P., Ella R., Deshpande G., Patil D. Y., Nyayanit D. A., Gupta N., Sahay R. R., Shete A. M., Panda S., Bhargava B., Mohan V. K., Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin. Infect. Dis. ciab411 (2021). 10.1093/cid/ciab41110.1093/cid/ciab411 - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

