Advances in the Development Ubiquitin-Specific Peptidase (USP) Inhibitors
- PMID: 33925279
- PMCID: PMC8123678
- DOI: 10.3390/ijms22094546
Advances in the Development Ubiquitin-Specific Peptidase (USP) Inhibitors
Abstract
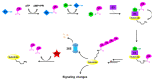
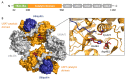
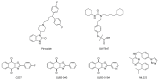
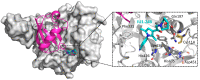
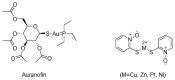
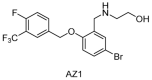
Ubiquitylation and deubiquitylation are reversible protein post-translational modification (PTM) processes involving the regulation of protein degradation under physiological conditions. Loss of balance in this regulatory system can lead to a wide range of diseases, such as cancer and inflammation. As the main members of the deubiquitinases (DUBs) family, ubiquitin-specific peptidases (USPs) are closely related to biological processes through a variety of molecular signaling pathways, including DNA damage repair, p53 and transforming growth factor-β (TGF-β) pathways. Over the past decade, increasing attention has been drawn to USPs as potential targets for the development of therapeutics across diverse therapeutic areas. In this review, we summarize the crucial roles of USPs in different signaling pathways and focus on advances in the development of USP inhibitors, as well as the methods of screening and identifying USP inhibitors.
Keywords: USP inhibitors; drug screening; signaling pathways; ubiquitin-specific peptidases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

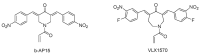
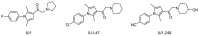
Similar articles
-
The role of ubiquitin-specific peptidases in cancer progression.J Biomed Sci. 2019 May 27;26(1):42. doi: 10.1186/s12929-019-0522-0. J Biomed Sci. 2019. PMID: 31133011 Free PMC article. Review.
-
Structure and function of the highly homologous deubiquitinases ubiquitin specific peptidase 25 and 28: Insights into their pathophysiological and therapeutic roles.Biochem Pharmacol. 2023 Jul;213:115624. doi: 10.1016/j.bcp.2023.115624. Epub 2023 May 26. Biochem Pharmacol. 2023. PMID: 37245535 Review.
-
Role of Ubiquitin-specific Proteases in Hepatocellular Carcinoma Pathogenesis.Curr Top Med Chem. 2024;24(3):179-191. doi: 10.2174/0115680266279228231219101233. Curr Top Med Chem. 2024. PMID: 38173207 Review.
-
Ubiquitin specific peptidases and prostate cancer.PeerJ. 2023 Feb 16;11:e14799. doi: 10.7717/peerj.14799. eCollection 2023. PeerJ. 2023. PMID: 36811009 Free PMC article. Review.
-
Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes.Pharmaceuticals (Basel). 2021 Aug 26;14(9):848. doi: 10.3390/ph14090848. Pharmaceuticals (Basel). 2021. PMID: 34577547 Free PMC article. Review.
Cited by
-
Deubiquitinases in Ovarian Cancer: Role in Drug Resistance and Tumor Aggressiveness.Int J Biol Sci. 2024 Sep 23;20(13):5208-5222. doi: 10.7150/ijbs.100355. eCollection 2024. Int J Biol Sci. 2024. PMID: 39430244 Free PMC article. Review.
-
Repurposing of US-FDA-approved drugs as negative modulators of ubiquitin specific protease-7 (USP7).Heliyon. 2024 Feb 23;10(5):e26345. doi: 10.1016/j.heliyon.2024.e26345. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38468948 Free PMC article.
-
The Ubiquitin-Proteasome System in Tumor Metabolism.Cancers (Basel). 2023 Apr 20;15(8):2385. doi: 10.3390/cancers15082385. Cancers (Basel). 2023. PMID: 37190313 Free PMC article. Review.
-
Artificial intelligence for prediction of biological activities and generation of molecular hits using stereochemical information.J Comput Aided Mol Des. 2023 Dec;37(12):791-806. doi: 10.1007/s10822-023-00539-9. Epub 2023 Oct 17. J Comput Aided Mol Des. 2023. PMID: 37847342 Free PMC article.
-
Unraveling the Immune Regulatory Functions of USP5: Implications for Disease Therapy.Biomolecules. 2024 Jun 12;14(6):683. doi: 10.3390/biom14060683. Biomolecules. 2024. PMID: 38927085 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

