The p23 of Citrus Tristeza Virus Interacts with Host FKBP-Type Peptidyl-Prolylcis-Trans Isomerase 17-2 and Is Involved in the Intracellular Movement of the Viral Coat Protein
- PMID: 33920690
- PMCID: PMC8073322
- DOI: 10.3390/cells10040934
The p23 of Citrus Tristeza Virus Interacts with Host FKBP-Type Peptidyl-Prolylcis-Trans Isomerase 17-2 and Is Involved in the Intracellular Movement of the Viral Coat Protein
Abstract
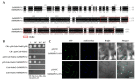
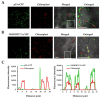
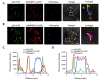
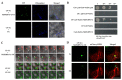
Citrus tristeza virus is a member of the genus Closterovirus in the family Closteroviridae. The p23 of citrus tristeza virus (CTV) is a multifunctional protein and RNA silencing suppressor. In this study, we identified a p23 interacting partner, FK506-binding protein (FKBP) 17-2, from Citrus aurantifolia (CaFKBP17-2), a susceptible host, and Nicotiana benthamiana (NbFKBP17-2), an experimental host for CTV. The interaction of p23 with CaFKBP17-2 and NbFKBP17-2 were individually confirmed by yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays. Subcellular localization tests showed that the viral p23 translocated FKBP17-2 from chloroplasts to the plasmodesmata of epidermal cells of N. benthamiana leaves. The knocked-down expression level of NbFKBP17-2 mRNA resulted in a decreased CTV titer in N. benthamiana plants. Further, BiFC and Y2H assays showed that NbFKBP17-2 also interacted with the coat protein (CP) of CTV, and the complexes of CP/NbFKBP17-2 rapidly moved in the cytoplasm. Moreover, p23 guided the CP/NbFKBP17-2 complexes to move along the cell wall. To the best of our knowledge, this is the first report of viral proteins interacting with FKBP17-2 encoded by plants. Our results provide insights for further revealing the mechanism of the CTV CP protein movement.
Keywords: FK506-binding protein; citrus tristeza virus; coat protein; p23; protein-protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Citrus tristeza virus co-opts glyceraldehyde 3-phosphate dehydrogenase for its infectious cycle by interacting with the viral-encoded protein p23.Plant Mol Biol. 2018 Nov;98(4-5):363-373. doi: 10.1007/s11103-018-0783-0. Epub 2018 Nov 3. Plant Mol Biol. 2018. PMID: 30392159 Free PMC article.
-
Citrus tristeza virus p23: determinants for nucleolar localization and their influence on suppression of RNA silencing and pathogenesis.Mol Plant Microbe Interact. 2013 Mar;26(3):306-18. doi: 10.1094/MPMI-08-12-0201-R. Mol Plant Microbe Interact. 2013. PMID: 23387469
-
The Coat Protein of Citrus Yellow Vein Clearing Virus Interacts with Viral Movement Proteins and Serves as an RNA Silencing Suppressor.Viruses. 2019 Apr 5;11(4):329. doi: 10.3390/v11040329. Viruses. 2019. PMID: 30959816 Free PMC article.
-
Citrus tristeza virus: a pathogen that changed the course of the citrus industry.Mol Plant Pathol. 2008 Mar;9(2):251-68. doi: 10.1111/j.1364-3703.2007.00455.x. Mol Plant Pathol. 2008. PMID: 18705856 Free PMC article. Review.
-
From the smallest to the largest subcellular plant pathogen: Citrus tristeza virus and its unique p23 protein.Virus Res. 2022 Jun;314:198755. doi: 10.1016/j.virusres.2022.198755. Epub 2022 Mar 24. Virus Res. 2022. PMID: 35341876 Review.
Cited by
-
Protein P5 of pear chlorotic leaf spot-associated virus is a pathogenic factor that suppresses RNA silencing and enhances virus movement.Mol Plant Pathol. 2024 Oct;25(10):e70015. doi: 10.1111/mpp.70015. Mol Plant Pathol. 2024. PMID: 39412447 Free PMC article.
-
Transcriptomic alterations in the sweet orange vasculature correlate with growth repression induced by a variant of citrus tristeza virus.Front Microbiol. 2023 Apr 17;14:1162613. doi: 10.3389/fmicb.2023.1162613. eCollection 2023. Front Microbiol. 2023. PMID: 37138615 Free PMC article.
-
Identification and Functional Analyses of Host Proteins Interacting with the P3a Protein of Brassica Yellows Virus.Biology (Basel). 2023 Jan 28;12(2):202. doi: 10.3390/biology12020202. Biology (Basel). 2023. PMID: 36829481 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

