Long Non-Coding RNAs in Insects
- PMID: 33919662
- PMCID: PMC8069800
- DOI: 10.3390/ani11041118
Long Non-Coding RNAs in Insects
Abstract
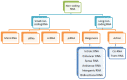
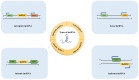
Only a small subset of all the transcribed RNAs are used as a template for protein translation, whereas RNA molecules that are not translated play a very important role as regulatory non-coding RNAs (ncRNAs). Besides traditionally known RNAs (ribosomal and transfer RNAs), ncRNAs also include small non-coding RNAs (sncRNAs) and long non-coding RNAs (lncRNAs). The lncRNAs, which were initially thought to be junk, have gained a great deal attention because of their regulatory roles in diverse biological processes in animals and plants. Insects are the most abundant and diverse group of animals on this planet. Recent studies have demonstrated the role of lncRNAs in almost all aspects of insect development, reproduction, and genetic plasticity. In this review, we describe the function and molecular mechanisms of the mode of action of different insect lncRNAs discovered up to date.
Keywords: LncRNAs; development; insects; non-coding RNA; regulatory functions.
Conflict of interest statement
The authors declare no conflict of interest whatsoever.
Figures


Similar articles
-
LncRNAs exert indispensable roles in orchestrating the interaction among diverse noncoding RNAs and enrich the regulatory network of plant growth and its adaptive environmental stress response.Hortic Res. 2023 Nov 17;10(12):uhad234. doi: 10.1093/hr/uhad234. eCollection 2023 Dec. Hortic Res. 2023. PMID: 38156284 Free PMC article.
-
Regulatory non-coding RNAs: a new frontier in regulation of plant biology.Funct Integr Genomics. 2021 Jul;21(3-4):313-330. doi: 10.1007/s10142-021-00787-8. Epub 2021 May 20. Funct Integr Genomics. 2021. PMID: 34013486 Free PMC article. Review.
-
The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring.Crit Rev Clin Lab Sci. 2014 Dec;51(6):344-57. doi: 10.3109/10408363.2014.944299. Epub 2014 Aug 15. Crit Rev Clin Lab Sci. 2014. PMID: 25123609 Review.
-
Regulatory non-coding RNAs: revolutionizing the RNA world.Mol Biol Rep. 2014 Jun;41(6):3915-23. doi: 10.1007/s11033-014-3259-6. Epub 2014 Feb 19. Mol Biol Rep. 2014. PMID: 24549720 Review.
-
Translation and emerging functions of non-coding RNAs in inflammation and immunity.Allergy. 2022 Jul;77(7):2025-2037. doi: 10.1111/all.15234. Epub 2022 Feb 9. Allergy. 2022. PMID: 35094406 Free PMC article. Review.
Cited by
-
Genome-wide analysis of long noncoding RNAs and their association in regulating the metamorphosis of the Sarcophaga peregrina (Diptera: Sarcophagidae).PLoS Negl Trop Dis. 2023 Jun 26;17(6):e0011411. doi: 10.1371/journal.pntd.0011411. eCollection 2023 Jun. PLoS Negl Trop Dis. 2023. PMID: 37363930 Free PMC article.
-
MicroRNA Targets PAP1 to Mediate Melanization in Plutella xylostella (Linnaeus) Infected by Metarhizium anisopliae.Int J Mol Sci. 2024 Jan 17;25(2):1140. doi: 10.3390/ijms25021140. Int J Mol Sci. 2024. PMID: 38256210 Free PMC article.
-
LncRNA and Protein Expression Profiles Reveal Heart Adaptation to High-Altitude Hypoxia in Tibetan Sheep.Int J Mol Sci. 2023 Dec 27;25(1):385. doi: 10.3390/ijms25010385. Int J Mol Sci. 2023. PMID: 38203557 Free PMC article.
-
Phenotypic Plasticity: What Has DNA Methylation Got to Do with It?Insects. 2022 Jan 19;13(2):110. doi: 10.3390/insects13020110. Insects. 2022. PMID: 35206684 Free PMC article. Review.
-
Editorial: Role of Non-Coding RNAs in Animals.Animals (Basel). 2023 Feb 23;13(5):805. doi: 10.3390/ani13050805. Animals (Basel). 2023. PMID: 36899662 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

