Protein Binding Partners of Dysregulated miRNAs in Parkinson's Disease Serum
- PMID: 33918274
- PMCID: PMC8065836
- DOI: 10.3390/cells10040791
Protein Binding Partners of Dysregulated miRNAs in Parkinson's Disease Serum
Abstract
Accumulating evidence suggests that microRNAs (miRNAs) are a contributing factor to neurodegenerative diseases. Although altered miRNA profiles in serum or plasma have been reported for several neurodegenerative diseases, little is known about the interaction between dysregulated miRNAs and their protein binding partners. We found significant alterations of the miRNA abundance pattern in serum and in isolated serum-derived extracellular vesicles of Parkinson's disease (PD) patients. The differential expression of miRNA in PD patients was more robust in serum than in isolated extracellular vesicles and could separate PD patients from healthy controls in an unsupervised approach to a high degree. We identified a novel protein interaction partner for the strongly dysregulated hsa-mir-4745-5p. Our study provides further evidence for the involvement of miRNAs and HNF4a in PD. The demonstration that miRNA-protein binding might mediate the pathologic effects of HNF4a both by direct binding to it and by binding to proteins regulated by it suggests a complex role for miRNAs in pathology beyond the dysregulation of transcription.
Keywords: Parkinson’s disease; miRNA; serum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
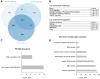
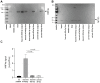
Similar articles
-
Several miRNAs derived from serum extracellular vesicles are potential biomarkers for early diagnosis and progression of Parkinson's disease.Transl Neurodegener. 2021 Jul 28;10(1):25. doi: 10.1186/s40035-021-00249-y. Transl Neurodegener. 2021. PMID: 34315539 Free PMC article.
-
Identification of plasma microRNA expression changes in multiple system atrophy and Parkinson's disease.Mol Brain. 2019 May 14;12(1):49. doi: 10.1186/s13041-019-0471-2. Mol Brain. 2019. PMID: 31088501 Free PMC article.
-
Exosomal miRNA as peripheral biomarkers in Parkinson's disease and progressive supranuclear palsy: A pilot study.Parkinsonism Relat Disord. 2021 Dec;93:77-84. doi: 10.1016/j.parkreldis.2021.11.020. Epub 2021 Nov 24. Parkinsonism Relat Disord. 2021. PMID: 34839044
-
Exosomal microRNAs in Parkinson's disease: insights into biomarker potential and disease pathology.Neurol Sci. 2024 Aug;45(8):3625-3639. doi: 10.1007/s10072-024-07439-2. Epub 2024 Mar 27. Neurol Sci. 2024. PMID: 38532190 Review.
-
microRNAs involved in Parkinson's disease: A systematic review.Mol Med Rep. 2016 Nov;14(5):4015-4022. doi: 10.3892/mmr.2016.5759. Epub 2016 Sep 22. Mol Med Rep. 2016. PMID: 27666518 Review.
Cited by
-
Epigenetic Changes in Prion and Prion-like Neurodegenerative Diseases: Recent Advances, Potential as Biomarkers, and Future Perspectives.Int J Mol Sci. 2022 Oct 20;23(20):12609. doi: 10.3390/ijms232012609. Int J Mol Sci. 2022. PMID: 36293477 Free PMC article. Review.
-
Common microRNA regulated pathways in Alzheimer's and Parkinson's disease.Front Neurosci. 2023 Sep 1;17:1228927. doi: 10.3389/fnins.2023.1228927. eCollection 2023. Front Neurosci. 2023. PMID: 37719162 Free PMC article.
-
Extracellular vesicles in the study of Alzheimer's and Parkinson's diseases: Methodologies applied from cells to biofluids.J Neurochem. 2022 Nov;163(4):266-309. doi: 10.1111/jnc.15697. Epub 2022 Oct 22. J Neurochem. 2022. PMID: 36156258 Free PMC article. Review.
-
Identification and characterization of virus-encoded circular RNAs in host cells.Microb Genom. 2022 Jun;8(6):mgen000848. doi: 10.1099/mgen.0.000848. Microb Genom. 2022. PMID: 35731570 Free PMC article.
-
The Fragile X Protein Family in Amyotrophic Lateral Sclerosis.Mol Neurobiol. 2023 Jul;60(7):3898-3910. doi: 10.1007/s12035-023-03330-x. Epub 2023 Mar 29. Mol Neurobiol. 2023. PMID: 36991279 Free PMC article. Review.
References
-
- Freischmidt A., Müller K., Zondler L., Weydt P., Volk A.E., Božič A.L., Walter M., Bonin M., Mayer B., von Arnim C.A.F., et al. Serum microRNAs in patients with genetic amyotrophic lateral sclerosis and pre-manifest mutation carriers. Brain. 2014;137:2938–2950. doi: 10.1093/brain/awu249. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

