Transcription-coupled nucleotide excision repair: New insights revealed by genomic approaches
- PMID: 33894524
- PMCID: PMC8205993
- DOI: 10.1016/j.dnarep.2021.103126
Transcription-coupled nucleotide excision repair: New insights revealed by genomic approaches
Abstract
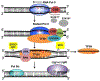
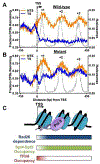
Elongation of RNA polymerase II (Pol II) is affected by many factors including DNA damage. Bulky damage, such as lesions caused by ultraviolet (UV) radiation, arrests Pol II and inhibits gene transcription, and may lead to genome instability and cell death. Cells activate transcription-coupled nucleotide excision repair (TC-NER) to remove Pol II-impeding damage and allow transcription resumption. TC-NER initiation in humans is mediated by Cockayne syndrome group B (CSB) protein, which binds to the stalled Pol II and promotes assembly of the repair machinery. Given the complex nature of the TC-NER pathway and its unique function at the interface between transcription and repair, new approaches are required to gain in-depth understanding of the mechanism. Advances in genomic approaches provide an important opportunity to investigate how TC-NER is initiated upon damage-induced Pol II stalling and what factors are involved in this process. In this Review, we discuss new mechanisms of TC-NER revealed by genome-wide DNA damage mapping and new TC-NER factors identified by high-throughput screening. As TC-NER conducts strand-specific repair of mutagenic damage, we also discuss how this repair pathway causes mutational strand asymmetry in the cancer genome.
Keywords: CPD-seq; CSB; DNA damage; Mutagenesis; RNA Pol II.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures

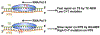
Similar articles
-
Differential processing of RNA polymerase II at DNA damage correlates with transcription-coupled repair syndrome severity.Nucleic Acids Res. 2024 Sep 9;52(16):9596-9612. doi: 10.1093/nar/gkae618. Nucleic Acids Res. 2024. PMID: 39021334 Free PMC article.
-
Structural basis of DNA lesion recognition for eukaryotic transcription-coupled nucleotide excision repair.DNA Repair (Amst). 2018 Nov;71:43-55. doi: 10.1016/j.dnarep.2018.08.006. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30174298 Free PMC article. Review.
-
DNA damage response and transcription.DNA Repair (Amst). 2011 Jul 15;10(7):743-50. doi: 10.1016/j.dnarep.2011.04.024. Epub 2011 May 31. DNA Repair (Amst). 2011. PMID: 21622031 Review.
-
What happens at the lesion does not stay at the lesion: Transcription-coupled nucleotide excision repair and the effects of DNA damage on transcription in cis and trans.DNA Repair (Amst). 2018 Nov;71:56-68. doi: 10.1016/j.dnarep.2018.08.007. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30195642 Review.
-
Elf1 promotes transcription-coupled repair in yeast by using its C-terminal domain to bind TFIIH.Nat Commun. 2024 Jul 23;15(1):6223. doi: 10.1038/s41467-024-50539-y. Nat Commun. 2024. PMID: 39043658 Free PMC article.
Cited by
-
Busulfan Chemotherapy Downregulates TAF7/TNF-α Signaling in Male Germ Cell Dysfunction.Biomedicines. 2024 Sep 28;12(10):2220. doi: 10.3390/biomedicines12102220. Biomedicines. 2024. PMID: 39457533 Free PMC article.
-
Genome-wide mapping of genomic DNA damage: methods and implications.Cell Mol Life Sci. 2021 Nov;78(21-22):6745-6762. doi: 10.1007/s00018-021-03923-6. Epub 2021 Aug 31. Cell Mol Life Sci. 2021. PMID: 34463773 Free PMC article. Review.
-
Do Noncoding and Coding Sites in Angiosperm Chloroplast DNA Have Different Mutation Processes?Genes (Basel). 2023 Jan 5;14(1):148. doi: 10.3390/genes14010148. Genes (Basel). 2023. PMID: 36672890 Free PMC article.
-
DNA damage and repair: underlying mechanisms leading to microcephaly.Front Cell Dev Biol. 2023 Oct 10;11:1268565. doi: 10.3389/fcell.2023.1268565. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37881689 Free PMC article. Review.
-
Susceptibility of DNA damage recognition activities linked to nucleotide excision and mismatch repair in zebrafish (Danio rerio) early and mid-early embryos to 2.5 to 4.5 °C heat stress.Fish Physiol Biochem. 2023 Jun;49(3):515-527. doi: 10.1007/s10695-023-01198-1. Epub 2023 May 3. Fish Physiol Biochem. 2023. PMID: 37133645
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

