Adhesion receptor ADGRG2/GPR64 is in the GI-tract selectively expressed in mature intestinal tuft cells
- PMID: 33831593
- PMCID: PMC8105302
- DOI: 10.1016/j.molmet.2021.101231
Adhesion receptor ADGRG2/GPR64 is in the GI-tract selectively expressed in mature intestinal tuft cells
Abstract
Objective: GPR64/ADGRG2 is an orphan Adhesion G protein-coupled receptor (ADGR) known to be mainly expressed in the parathyroid gland and epididymis. This investigation aimed to delineate the cellular expression of GPR64 throughout the body with focus on the gastrointestinal (GI) tract.
Methods: Transgenic Gpr64mCherry reporter mice were histologically examined throughout the body and reporter protein expression in intestinal tuft cells was confirmed by specific cell ablation. The GPCR repertoire of intestinal Gpr64mCherry-positive tuft cells was analyzed by quantitative RT-PCR analysis and in situ hybridization. The Gpr64mCherry was crossed into the general tuft cell reporter Trpm5GFP to generate small intestinal organoids for time-lapse imaging. Intestinal tuft cells were isolated from small intestine, FACS-purified and transcriptionally compared using RNA-seq analysis.
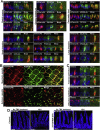
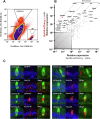
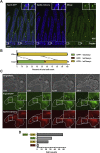
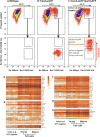
Results: Expression of the Gpr64mCherry reporter was identified in multiple organs and specifically in olfactory microvillous cells, enteric nerves, and importantly in respiratory and GI tuft cells. In the small intestine, cell ablation targeting Gpr64-expressing epithelial cells eliminated tuft cells. Transcriptional analysis of small intestinal Gpr64mCherry -positive tuft cells confirmed expression of Gpr64 and the chemo-sensors Sucnr1, Gprc5c, Drd3, and Gpr41/Ffar3. Time-lapse studies of organoids from Trpm5GFP:Gpr64mCherry mice revealed sequential expression of initially Trpm5GFP and subsequently also Gpr64mCherry in maturing intestinal tuft cells. RNA-seq analysis of small intestinal tuft cells based on these two markers demonstrated a dynamic change in expression of transcription factors and GPCRs from young to mature tuft cells.
Conclusions: GPR64 is expressed in chemosensory epithelial cells across a broad range of tissues; however, in the GI tract, GPR64 is remarkably selectively expressed in mature versus young immunoregulatory tuft cells.
Keywords: ADGRG2; Chemosensory cells; GPCRs; GPR64; Tuft cells.
Copyright © 2021 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures

Similar articles
-
Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine.Proc Natl Acad Sci U S A. 2018 May 22;115(21):5552-5557. doi: 10.1073/pnas.1720758115. Epub 2018 May 7. Proc Natl Acad Sci U S A. 2018. PMID: 29735652 Free PMC article.
-
Orphan Adhesion GPCR GPR64/ADGRG2 Is Overexpressed in Parathyroid Tumors and Attenuates Calcium-Sensing Receptor-Mediated Signaling.J Bone Miner Res. 2017 Mar;32(3):654-666. doi: 10.1002/jbmr.3023. Epub 2016 Nov 25. J Bone Miner Res. 2017. PMID: 27760455 Free PMC article.
-
Advillin is a tuft cell marker in the mouse alimentary tract.J Mol Histol. 2020 Aug;51(4):421-435. doi: 10.1007/s10735-020-09893-6. Epub 2020 Jul 2. J Mol Histol. 2020. PMID: 32617896 Free PMC article.
-
Luminal chemosensing in the gastroduodenal mucosa.Curr Opin Gastroenterol. 2017 Nov;33(6):439-445. doi: 10.1097/MOG.0000000000000396. Curr Opin Gastroenterol. 2017. PMID: 28806271 Free PMC article. Review.
-
Dclk1-expressing tuft cells: critical modulators of the intestinal niche?Am J Physiol Gastrointest Liver Physiol. 2017 Oct 1;313(4):G285-G299. doi: 10.1152/ajpgi.00073.2017. Epub 2017 Jul 6. Am J Physiol Gastrointest Liver Physiol. 2017. PMID: 28684459 Free PMC article. Review.
Cited by
-
Enteroendocrine Cell Formation Is an Early Event in Pancreatic Tumorigenesis.Front Physiol. 2022 Apr 27;13:865452. doi: 10.3389/fphys.2022.865452. eCollection 2022. Front Physiol. 2022. PMID: 35574446 Free PMC article.
-
Colonic Tuft Cells: The Less-Recognized Therapeutic Targets in Inflammatory Bowel Disease and Colorectal Cancer.Int J Mol Sci. 2024 Jun 5;25(11):6209. doi: 10.3390/ijms25116209. Int J Mol Sci. 2024. PMID: 38892399 Free PMC article. Review.
-
G-protein coupled receptor 5C (GPRC5C) is required for osteoblast differentiation and responds to EZH2 inhibition and multiple osteogenic signals.Bone. 2023 Nov;176:116866. doi: 10.1016/j.bone.2023.116866. Epub 2023 Aug 7. Bone. 2023. PMID: 37558192 Free PMC article.
-
Proteomic approach towards identification of seminal fluid biomarkers from individuals with severe oligozoospermia, cryptozoospermia and non-obstructive azoospermia: a pilot study.Transl Androl Urol. 2023 Oct 31;12(10):1497-1510. doi: 10.21037/tau-23-130. Epub 2023 Oct 24. Transl Androl Urol. 2023. PMID: 37969768 Free PMC article.
-
Tuft Cells and Their Role in Intestinal Diseases.Front Immunol. 2022 Feb 14;13:822867. doi: 10.3389/fimmu.2022.822867. eCollection 2022. Front Immunol. 2022. PMID: 35237268 Free PMC article. Review.
References
-
- Crosnier C., Stamataki D., Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nature Reviews Genetics. 2006;7(5):349–359. - PubMed
-
- Engelstoft M.S., Egerod K.L., Holst B., Schwartz T.W. A gut feeling for obesity: 7TM sensors on enteroendocrine cells. Cell Metabolism. 2008;8(6):447–449. - PubMed
-
- Rhodin J., Dalhamn T. Electron microscopy of the tracheal ciliated mucosa in rat. Zeitschrift fur Zellforschung und mikroskopische Anatomie. 1956;44(4):345–412. - PubMed
-
- Chang L.Y., Mercer R.R., Crapo J.D. Differential distribution of brush cells in the rat lung. The Anatomical Record. 1986;216(1):49–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

