Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite
- PMID: 33789084
- PMCID: PMC7953435
- DOI: 10.1016/j.chom.2021.03.005
Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite
Abstract
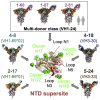
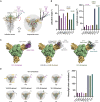
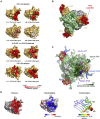
Numerous antibodies that neutralize SARS-CoV-2 have been identified, and these generally target either the receptor-binding domain (RBD) or the N-terminal domain (NTD) of the viral spike. While RBD-directed antibodies have been extensively studied, far less is known about NTD-directed antibodies. Here, we report cryo-EM and crystal structures for seven potent NTD-directed neutralizing antibodies in complex with spike or isolated NTD. These structures defined several antibody classes, with at least one observed in multiple convalescent donors. The structures revealed that all seven antibodies target a common surface, bordered by glycans N17, N74, N122, and N149. This site-formed primarily by a mobile β-hairpin and several flexible loops-was highly electropositive, located at the periphery of the spike, and the largest glycan-free surface of NTD facing away from the viral membrane. Thus, in contrast to neutralizing RBD-directed antibodies that recognize multiple non-overlapping epitopes, potent NTD-directed neutralizing antibodies appear to target a single supersite.
Keywords: COVID-19; N-terminal domain; SARS-CoV-2; antibody class; antigenic supersite; multi-donor antibody; neutralizing antibody.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.D.H., Y.H., J.Y., L.L., M.S.N., and P.W. are inventors of a patent describing some of the antibodies reported on here.
Figures





Comment in
-
An NTD supersite of attack.Cell Host Microbe. 2021 May 12;29(5):744-746. doi: 10.1016/j.chom.2021.04.010. Cell Host Microbe. 2021. PMID: 33984277 Free PMC article.
Similar articles
-
Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant.Immunity. 2021 Jun 8;54(6):1276-1289.e6. doi: 10.1016/j.immuni.2021.03.023. Epub 2021 Apr 1. Immunity. 2021. PMID: 33836142 Free PMC article.
-
Limited Variation between SARS-CoV-2-Infected Individuals in Domain Specificity and Relative Potency of the Antibody Response against the Spike Glycoprotein.Microbiol Spectr. 2022 Feb 23;10(1):e0267621. doi: 10.1128/spectrum.02676-21. Epub 2022 Jan 26. Microbiol Spectr. 2022. PMID: 35080430 Free PMC article.
-
Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.Nature. 2020 Aug;584(7821):450-456. doi: 10.1038/s41586-020-2571-7. Epub 2020 Jul 22. Nature. 2020. PMID: 32698192
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
-
Analysis of the molecular mechanism of SARS-CoV-2 antibodies.Biochem Biophys Res Commun. 2021 Aug 20;566:45-52. doi: 10.1016/j.bbrc.2021.06.001. Epub 2021 Jun 5. Biochem Biophys Res Commun. 2021. PMID: 34116356 Free PMC article. Review.
Cited by
-
Biophysical and structural characterizations of the effects of mutations on the structure-activity relationships of SARS-CoV-2 spike protein.Methods Enzymol. 2022;675:299-321. doi: 10.1016/bs.mie.2022.07.013. Epub 2022 Aug 19. Methods Enzymol. 2022. PMID: 36220274 Free PMC article.
-
An NTD supersite of attack.Cell Host Microbe. 2021 May 12;29(5):744-746. doi: 10.1016/j.chom.2021.04.010. Cell Host Microbe. 2021. PMID: 33984277 Free PMC article.
-
Antibodies to S2 domain of SARS-CoV-2 spike protein in Moderna mRNA vaccinated subjects sustain antibody-dependent NK cell-mediated cell cytotoxicity against Omicron BA.1.Front Immunol. 2023 Nov 21;14:1266829. doi: 10.3389/fimmu.2023.1266829. eCollection 2023. Front Immunol. 2023. PMID: 38077368 Free PMC article.
-
Differential laboratory passaging of SARS-CoV-2 viral stocks impacts the in vitro assessment of neutralizing antibodies.PLoS One. 2024 Jan 25;19(1):e0289198. doi: 10.1371/journal.pone.0289198. eCollection 2024. PLoS One. 2024. PMID: 38271318 Free PMC article.
-
The impact of high-resolution structural data on stemming the COVID-19 pandemic.Curr Opin Virol. 2021 Aug;49:127-138. doi: 10.1016/j.coviro.2021.05.005. Epub 2021 Jun 3. Curr Opin Virol. 2021. PMID: 34130040 Free PMC article. Review.
References
-
- Adams P.D., Gopal K., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Pai R.K., Read R.J., Romo T.D. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 2004;11:53–55. - PubMed
-
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

