In silico analysis of altered expression of long non-coding RNA in SARS-CoV-2 infected cells and their possible regulation by STAT1, STAT3 and interferon regulatory factors
- PMID: 33688586
- PMCID: PMC7914022
- DOI: 10.1016/j.heliyon.2021.e06395
In silico analysis of altered expression of long non-coding RNA in SARS-CoV-2 infected cells and their possible regulation by STAT1, STAT3 and interferon regulatory factors
Abstract
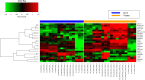
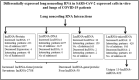
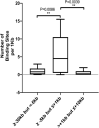
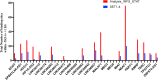
Altered expression of long noncoding RNA (lncRNA), longer than 200 nucleotides without potential for coding protein, has been observed in diverse human diseases including viral diseases. It is largely unknown whether lncRNA would deregulate in SARS-CoV-2 infection, causing ongoing pandemic COVID-19. To identify, if lncRNA was deregulated in SARS-CoV-2 infected cells, we analyzed in silico the data in GSE147507. It was revealed that expression of 20 lncRNA like MALAT1, NEAT1 was increased and 4 lncRNA like PART1, TP53TG1 was decreased in at least two independent cell lines infected with SARS-CoV-2. Expression of NEAT1 was also increased in lungs tissue of COVID-19 patients. The deregulated lncRNA could interact with more than 2800 genes/proteins and 422 microRNAs as revealed from the database that catalogs experimentally determined interactions. Analysis with the interacting gene/protein partners of deregulated lncRNAs revealed that these genes/proteins were associated with many pathways related to viral infection, inflammation and immune functions. To find out whether these lncRNAs could be regulated by STATs and interferon regulatory factors (IRFs), we used ChIPBase v2.0 that catalogs experimentally determined binding from ChIP-seq data. It was revealed that any one of the transcription factors IRF1, IRF4, STAT1, STAT3 and STAT5A had experimentally determined binding at regions within -5kb to +1kb of the deregulated lncRNAs in at least 2 independent cell lines/conditions. Our analysis revealed that several lncRNAs could be regulated by IRF1, IRF4 STAT1 and STAT3 in response to SARS-CoV-2 infection and lncRNAs might be involved in antiviral response. However, these in silico observations are necessary to be validated experimentally.
Keywords: COVID-19; Interferon regulatory factors; Long non-coding RNA; MALAT1; NEAT1; SARS-CoV-2.
© 2021 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Co-Regulation of Protein Coding Genes by Transcription Factor and Long Non-Coding RNA in SARS-CoV-2 Infected Cells: An In Silico Analysis.Noncoding RNA. 2021 Nov 29;7(4):74. doi: 10.3390/ncrna7040074. Noncoding RNA. 2021. PMID: 34940755 Free PMC article.
-
Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response.Genes (Basel). 2020 Jul 7;11(7):760. doi: 10.3390/genes11070760. Genes (Basel). 2020. PMID: 32646047 Free PMC article.
-
The interplay between lncRNAs, RNA-binding proteins and viral genome during SARS-CoV-2 infection reveals strong connections with regulatory events involved in RNA metabolism and immune response.Theranostics. 2022 May 9;12(8):3946-3962. doi: 10.7150/thno.73268. eCollection 2022. Theranostics. 2022. PMID: 35664076 Free PMC article.
-
The regulation of lncRNAs and miRNAs in SARS-CoV-2 infection.Front Cell Dev Biol. 2023 Jul 27;11:1229393. doi: 10.3389/fcell.2023.1229393. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37576600 Free PMC article. Review.
-
Long noncoding RNAs: Novel insights into hepatocelluar carcinoma.Cancer Lett. 2014 Mar 1;344(1):20-27. doi: 10.1016/j.canlet.2013.10.021. Epub 2013 Oct 30. Cancer Lett. 2014. PMID: 24183851 Review.
Cited by
-
Emerging Role of Interferon-Induced Noncoding RNA in Innate Antiviral Immunity.Viruses. 2022 Nov 23;14(12):2607. doi: 10.3390/v14122607. Viruses. 2022. PMID: 36560611 Free PMC article. Review.
-
Systems biology models to identify the influence of SARS-CoV-2 infections to the progression of human autoimmune diseases.Inform Med Unlocked. 2022;32:101003. doi: 10.1016/j.imu.2022.101003. Epub 2022 Jul 6. Inform Med Unlocked. 2022. PMID: 35818398 Free PMC article.
-
Long non-coding RNAs (lncRNAs) NEAT1 and MALAT1 are differentially expressed in severe COVID-19 patients: An integrated single-cell analysis.PLoS One. 2022 Jan 10;17(1):e0261242. doi: 10.1371/journal.pone.0261242. eCollection 2022. PLoS One. 2022. PMID: 35007307 Free PMC article.
-
COVID-19: Mechanisms, risk factors, genetics, non-coding RNAs and neurologic impairments.Noncoding RNA Res. 2023 Jun;8(2):240-254. doi: 10.1016/j.ncrna.2023.02.007. Epub 2023 Feb 23. Noncoding RNA Res. 2023. PMID: 36852336 Free PMC article. Review.
-
Evaluation of the Expression of Infection-Related Long Noncoding RNAs among COVID-19 Patients: A Case-Control Study.Genet Res (Camb). 2024 Feb 15;2024:3391054. doi: 10.1155/2024/3391054. eCollection 2024. Genet Res (Camb). 2024. PMID: 38389521 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

