De novo structural mutation rates and gamete-of-origin biases revealed through genome sequencing of 2,396 families
- PMID: 33675682
- PMCID: PMC8059337
- DOI: 10.1016/j.ajhg.2021.02.012
De novo structural mutation rates and gamete-of-origin biases revealed through genome sequencing of 2,396 families
Abstract
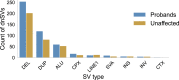
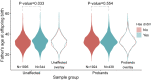
Each human genome includes de novo mutations that arose during gametogenesis. While these germline mutations represent a fundamental source of new genetic diversity, they can also create deleterious alleles that impact fitness. Whereas the rate and patterns of point mutations in the human germline are now well understood, far less is known about the frequency and features that impact de novo structural variants (dnSVs). We report a family-based study of germline mutations among 9,599 human genomes from 33 multigenerational CEPH-Utah families and 2,384 families from the Simons Foundation Autism Research Initiative. We find that de novo structural mutations detected by alignment-based, short-read WGS occur at an overall rate of at least 0.160 events per genome in unaffected individuals, and we observe a significantly higher rate (0.206 per genome) in ASD-affected individuals. In both probands and unaffected samples, nearly 73% of de novo structural mutations arose in paternal gametes, and we predict most de novo structural mutations to be caused by mutational mechanisms that do not require sequence homology. After multiple testing correction, we did not observe a statistically significant correlation between parental age and the rate of de novo structural variation in offspring. These results highlight that a spectrum of mutational mechanisms contribute to germline structural mutations and that these mechanisms most likely have markedly different rates and selective pressures than those leading to point mutations.
Keywords: autism; copy number variation; de novo mutation; genetic diversity; genomic structure; genomics; germline mutation; structural variation.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

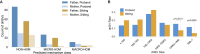
Similar articles
-
Identification and characterisation of de novo germline structural variants in two commercial pig lines using trio-based whole genome sequencing.BMC Genomics. 2023 Apr 18;24(1):208. doi: 10.1186/s12864-023-09296-3. BMC Genomics. 2023. PMID: 37072725 Free PMC article.
-
Parental influence on human germline de novo mutations in 1,548 trios from Iceland.Nature. 2017 Sep 28;549(7673):519-522. doi: 10.1038/nature24018. Epub 2017 Sep 20. Nature. 2017. PMID: 28959963
-
Patterns of de novo tandem repeat mutations and their role in autism.Nature. 2021 Jan;589(7841):246-250. doi: 10.1038/s41586-020-03078-7. Epub 2021 Jan 13. Nature. 2021. PMID: 33442040 Free PMC article.
-
Point mutations as a source of de novo genetic disease.Curr Opin Genet Dev. 2013 Jun;23(3):257-63. doi: 10.1016/j.gde.2013.01.007. Epub 2013 Mar 1. Curr Opin Genet Dev. 2013. PMID: 23453690 Review.
-
Properties and rates of germline mutations in humans.Trends Genet. 2013 Oct;29(10):575-84. doi: 10.1016/j.tig.2013.04.005. Epub 2013 May 16. Trends Genet. 2013. PMID: 23684843 Free PMC article. Review.
Cited by
-
MicroRNAs as Regulators, Biomarkers, and Therapeutic Targets in Autism Spectrum Disorder.Mol Neurobiol. 2024 Nov 6. doi: 10.1007/s12035-024-04582-x. Online ahead of print. Mol Neurobiol. 2024. PMID: 39503812 Review.
-
Jasmine and Iris: population-scale structural variant comparison and analysis.Nat Methods. 2023 Mar;20(3):408-417. doi: 10.1038/s41592-022-01753-3. Epub 2023 Jan 19. Nat Methods. 2023. PMID: 36658279 Free PMC article.
-
R-loop landscape in mature human sperm: Regulatory and evolutionary implications.Front Genet. 2023 Apr 17;14:1069871. doi: 10.3389/fgene.2023.1069871. eCollection 2023. Front Genet. 2023. PMID: 37139234 Free PMC article.
-
The multiple de novo copy number variant (MdnCNV) phenomenon presents with peri-zygotic DNA mutational signatures and multilocus pathogenic variation.Genome Med. 2022 Oct 27;14(1):122. doi: 10.1186/s13073-022-01123-w. Genome Med. 2022. PMID: 36303224 Free PMC article.
-
Genome sequencing and comprehensive rare-variant analysis of 465 families with neurodevelopmental disorders.Am J Hum Genet. 2023 Aug 3;110(8):1343-1355. doi: 10.1016/j.ajhg.2023.07.007. Am J Hum Genet. 2023. PMID: 37541188 Free PMC article.
References
-
- Halliday J.A., Glickman B.W. Mechanisms of spontaneous mutation in DNA repair-proficient Escherichia coli. Mutat. Res. 1991;250:55–71. - PubMed
-
- Friedberg E.C. DNA damage and repair. Nature. 2003;421:436–440. - PubMed
-
- Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

