The origin and early spread of SARS-CoV-2 in Europe
- PMID: 33571105
- PMCID: PMC7936359
- DOI: 10.1073/pnas.2012008118
The origin and early spread of SARS-CoV-2 in Europe
Abstract
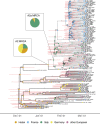
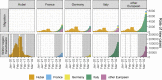
The investigation of migratory patterns during the SARS-CoV-2 pandemic before spring 2020 border closures in Europe is a crucial first step toward an in-depth evaluation of border closure policies. Here we analyze viral genome sequences using a phylodynamic model with geographic structure to estimate the origin and spread of SARS-CoV-2 in Europe prior to border closures. Based on SARS-CoV-2 genomes, we reconstruct a partial transmission tree of the early pandemic and coinfer the geographic location of ancestral lineages as well as the number of migration events into and between European regions. We find that the predominant lineage spreading in Europe during this time has a most recent common ancestor in Italy and was probably seeded by a transmission event in either Hubei, China or Germany. We do not find evidence for preferential migration paths from Hubei into different European regions or from each European region to the others. Sustained local transmission is first evident in Italy and then shortly thereafter in the other European regions considered. Before the first border closures in Europe, we estimate that the rate of occurrence of new cases from within-country transmission was within the bounds of the estimated rate of new cases from migration. In summary, our analysis offers a view on the early state of the epidemic in Europe and on migration patterns of the virus before border closures. This information will enable further study of the necessity and timeliness of border closures.
Keywords: SARS-CoV-2; disease import; phylogeography; transmission.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
Phylogeography and genomic epidemiology of SARS-CoV-2 in Italy and Europe with newly characterized Italian genomes between February-June 2020.Sci Rep. 2022 Apr 6;12(1):5736. doi: 10.1038/s41598-022-09738-0. Sci Rep. 2022. PMID: 35388091 Free PMC article.
-
A small number of early introductions seeded widespread transmission of SARS-CoV-2 in Québec, Canada.Genome Med. 2021 Oct 28;13(1):169. doi: 10.1186/s13073-021-00986-9. Genome Med. 2021. PMID: 34706766 Free PMC article.
-
Phylogeography of SARS-CoV-2 pandemic in Spain: a story of multiple introductions, micro-geographic stratification, founder effects, and super-spreaders.Zool Res. 2020 Nov 18;41(6):605-620. doi: 10.24272/j.issn.2095-8137.2020.217. Zool Res. 2020. PMID: 32935498 Free PMC article.
-
Did border closures slow SARS-CoV-2?Sci Rep. 2022 Feb 1;12(1):1709. doi: 10.1038/s41598-022-05482-7. Sci Rep. 2022. PMID: 35105912 Free PMC article. Review.
-
Topological Analysis for Sequence Variability: Case Study on more than 2K SARS-CoV-2 sequences of COVID-19 infected 54 countries in comparison with SARS-CoV-1 and MERS-CoV.Infect Genet Evol. 2021 Mar;88:104708. doi: 10.1016/j.meegid.2021.104708. Epub 2021 Jan 6. Infect Genet Evol. 2021. PMID: 33421654 Free PMC article. Review.
Cited by
-
Regional importation and asymmetric within-country spread of SARS-CoV-2 variants of concern in the Netherlands.medRxiv [Preprint]. 2022 Mar 22:2022.03.21.22272611. doi: 10.1101/2022.03.21.22272611. medRxiv. 2022. Update in: Elife. 2022 Sep 13;11:e78770. doi: 10.7554/eLife.78770. PMID: 35350194 Free PMC article. Updated. Preprint.
-
Genomic epidemiology of SARS-CoV-2 in a UK university identifies dynamics of transmission.Nat Commun. 2022 Feb 8;13(1):751. doi: 10.1038/s41467-021-27942-w. Nat Commun. 2022. PMID: 35136068 Free PMC article.
-
Cryptic transmission of SARS-CoV-2 and the first COVID-19 wave.Nature. 2021 Dec;600(7887):127-132. doi: 10.1038/s41586-021-04130-w. Epub 2021 Oct 25. Nature. 2021. PMID: 34695837 Free PMC article.
-
Tracking the onset date of the community spread of SARS-CoV-2 in western countries.Mem Inst Oswaldo Cruz. 2020;115:e200183. doi: 10.1590/0074-02760200183. Epub 2020 Sep 4. Mem Inst Oswaldo Cruz. 2020. PMID: 32901696 Free PMC article.
-
New Phylogenetic Models Incorporating Interval-Specific Dispersal Dynamics Improve Inference of Disease Spread.Mol Biol Evol. 2022 Aug 3;39(8):msac159. doi: 10.1093/molbev/msac159. Mol Biol Evol. 2022. PMID: 35861314 Free PMC article.
References
-
- Connor P., More than nine-in-ten people worldwide live in countries with travel restrictions amid COVID-19. Pew Research Center, https://www.pewresearch.org/fact-tank/2020/04/01/more-than-nine-in-ten-p.... Accessed 18 May 2020.
-
- WHO , Updated WHO recommendations for international traffic in relation to COVID-19 outbreak, https://www.who.int/news-room/articles-detail/updated-who-recommendation.... Accessed 18 May 2020.
-
- WHO , COVID-19 situation reports, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio.... Accessed 20 April 2020.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

