Regulation of host and virus genes by neuronal miR-138 favours herpes simplex virus 1 latency
- PMID: 33558653
- PMCID: PMC8221016
- DOI: 10.1038/s41564-020-00860-1
Regulation of host and virus genes by neuronal miR-138 favours herpes simplex virus 1 latency
Abstract
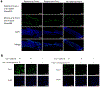
MicroRNA miR-138, which is highly expressed in neurons, represses herpes simplex virus 1 (HSV-1) lytic cycle genes by targeting viral ICP0 messenger RNA, thereby promoting viral latency in mice. We found that overexpressed miR-138 also represses lytic processes independently of ICP0 in murine and human neuronal cells; therefore, we investigated whether miR-138 has targets besides ICP0. Using genome-wide RNA sequencing/photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation followed by short interfering RNA knockdown of candidate targets, we identified the host Oct-1 and Foxc1 messenger mRNAs as miR-138's targets, whose gene products are transcription factors important for HSV-1 replication in neuronal cells. OCT-1 has a known role in the initiation of HSV transcription. Overexpression of FOXC1, which was not known to affect HSV-1, promoted HSV-1 replication in murine neurons and ganglia. CRISPR-Cas9 knockout of FOXC1 reduced viral replication, lytic gene expression and miR-138 repression in murine neuronal cells. FOXC1 also collaborated with ICP0 to decrease heterochromatin on viral genes and compensated for the defect of an ICP0-null virus. In summary, miR-138 targets ICP0, Oct-1 and Foxc1 to repress HSV-1 lytic cycle genes and promote epigenetic gene silencing, which together enable favourable conditions for latent infection.
Conflict of interest statement
Conflict of Interest
A.H.M.N. and G.M.C. are inventors on patents filed by the Presidents and Fellows of Harvard College. Full disclosure of G.M.C. is available on
Figures
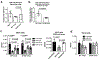






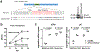
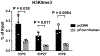





Similar articles
-
Neuronal miR-138 Represses HSV-2 Lytic Infection by Regulating Viral and Host Genes with Mechanistic Differences from HSV-1.J Virol. 2022 May 11;96(9):e0034922. doi: 10.1128/jvi.00349-22. Epub 2022 Apr 11. J Virol. 2022. PMID: 35404085 Free PMC article.
-
Mutations Inactivating Herpes Simplex Virus 1 MicroRNA miR-H2 Do Not Detectably Increase ICP0 Gene Expression in Infected Cultured Cells or Mouse Trigeminal Ganglia.J Virol. 2017 Jan 3;91(2):e02001-16. doi: 10.1128/JVI.02001-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847363 Free PMC article.
-
Increased neurovirulence and reactivation of the herpes simplex virus type 1 latency-associated transcript (LAT)-negative mutant dLAT2903 with a disrupted LAT miR-H2.J Neurovirol. 2016 Feb;22(1):38-49. doi: 10.1007/s13365-015-0362-y. Epub 2015 Jun 12. J Neurovirol. 2016. PMID: 26069184 Free PMC article.
-
The potential link between PML NBs and ICP0 in regulating lytic and latent infection of HSV-1.Protein Cell. 2012 May;3(5):372-82. doi: 10.1007/s13238-012-2021-x. Epub 2012 Apr 28. Protein Cell. 2012. PMID: 22544561 Free PMC article. Review.
-
ICP0, a regulator of herpes simplex virus during lytic and latent infection.Bioessays. 2000 Aug;22(8):761-70. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A. Bioessays. 2000. PMID: 10918307 Review.
Cited by
-
Small RNA sequencing analysis reveals regulation of microRNA expression in Madin-Darby canine kidney epithelial cells infected with Canid alphaherpesvirus 1.Virus Genes. 2024 Oct;60(5):537-548. doi: 10.1007/s11262-024-02091-6. Epub 2024 Jul 17. Virus Genes. 2024. PMID: 39017941
-
A review of HSV pathogenesis, vaccine development, and advanced applications.Mol Biomed. 2024 Aug 29;5(1):35. doi: 10.1186/s43556-024-00199-7. Mol Biomed. 2024. PMID: 39207577 Free PMC article. Review.
-
E3 ligase RNF5 inhibits type I interferon response in herpes simplex virus keratitis through the STING/IRF3 signaling pathway.Front Microbiol. 2022 Aug 2;13:944101. doi: 10.3389/fmicb.2022.944101. eCollection 2022. Front Microbiol. 2022. PMID: 35992663 Free PMC article.
-
Liver X Receptor-Inducible Host E3 Ligase IDOL Targets a Human Cytomegalovirus Reactivation Determinant.J Virol. 2023 Jul 27;97(7):e0075823. doi: 10.1128/jvi.00758-23. Epub 2023 Jun 20. J Virol. 2023. PMID: 37338407 Free PMC article.
-
Keeping division on track.Nat Microbiol. 2021 May;6(5):538-539. doi: 10.1038/s41564-021-00891-2. Nat Microbiol. 2021. PMID: 33927383 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

