Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases
- PMID: 33521695
- PMCID: PMC7837622
- DOI: 10.1016/j.xcrm.2021.100204
Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases
Abstract
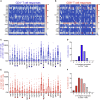
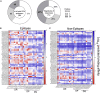
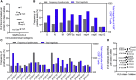
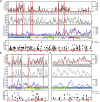
T cells are involved in control of SARS-CoV-2 infection. To establish the patterns of immunodominance of different SARS-CoV-2 antigens and precisely measure virus-specific CD4+ and CD8+ T cells, we study epitope-specific T cell responses of 99 convalescent coronavirus disease 2019 (COVID-19) cases. The SARS-CoV-2 proteome is probed using 1,925 peptides spanning the entire genome, ensuring an unbiased coverage of human leukocyte antigen (HLA) alleles for class II responses. For HLA class I, we study an additional 5,600 predicted binding epitopes for 28 prominent HLA class I alleles, accounting for wide global coverage. We identify several hundred HLA-restricted SARS-CoV-2-derived epitopes. Distinct patterns of immunodominance are observed, which differ for CD4+ T cells, CD8+ T cells, and antibodies. The class I and class II epitopes are combined into epitope megapools to facilitate identification and quantification of SARS-CoV-2-specific CD4+ and CD8+ T cells.
Keywords: CD4+T cells; CD8+ T cells; COVID-19; HLA; SARS-CoV-2; T cells; epitopes.
© 2021 The Author(s).
Conflict of interest statement
A.S. is a consultant for Gritstone, Flow Pharma, Merck, Epitogenesis, Gilead, and Avalia. S.C. is a consultant for Avalia. All other authors declare no competing interests. LJI has filed for patent protection for various aspects of vaccine design and identification of specific epitopes.
Figures



Update of
-
Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases.bioRxiv [Preprint]. 2020 Dec 9:2020.12.08.416750. doi: 10.1101/2020.12.08.416750. bioRxiv. 2020. Update in: Cell Rep Med. 2021 Feb 16;2(2):100204. doi: 10.1016/j.xcrm.2021.100204 PMID: 33330869 Free PMC article. Updated. Preprint.
Similar articles
-
Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases.bioRxiv [Preprint]. 2020 Dec 9:2020.12.08.416750. doi: 10.1101/2020.12.08.416750. bioRxiv. 2020. Update in: Cell Rep Med. 2021 Feb 16;2(2):100204. doi: 10.1016/j.xcrm.2021.100204 PMID: 33330869 Free PMC article. Updated. Preprint.
-
Parallel detection of SARS-CoV-2 epitopes reveals dynamic immunodominance profiles of CD8+ T memory cells in convalescent COVID-19 donors.Clin Transl Immunology. 2022 Oct 14;11(10):e1423. doi: 10.1002/cti2.1423. eCollection 2022. Clin Transl Immunology. 2022. PMID: 36254196 Free PMC article.
-
COVID-19 coronavirus vaccine T cell epitope prediction analysis based on distributions of HLA class I loci (HLA-A, -B, -C) across global populations.Hum Vaccin Immunother. 2021 Apr 3;17(4):1097-1108. doi: 10.1080/21645515.2020.1823777. Epub 2020 Nov 11. Hum Vaccin Immunother. 2021. PMID: 33175614 Free PMC article.
-
Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country.Front Immunol. 2020 Sep 3;11:2008. doi: 10.3389/fimmu.2020.02008. eCollection 2020. Front Immunol. 2020. PMID: 33013857 Free PMC article. Review.
-
Development of multi-epitope peptide-based vaccines against SARS-CoV-2.Biomed J. 2021 Mar;44(1):18-30. doi: 10.1016/j.bj.2020.09.005. Epub 2020 Oct 1. Biomed J. 2021. PMID: 33727051 Free PMC article. Review.
Cited by
-
In-silico evaluation of the T-cell based immune response against SARS-CoV-2 omicron variants.Sci Rep. 2024 Oct 25;14(1):25413. doi: 10.1038/s41598-024-75658-w. Sci Rep. 2024. PMID: 39455652 Free PMC article.
-
On the Origins of Omicron's Unique Spike Gene Insertion.Vaccines (Basel). 2022 Sep 9;10(9):1509. doi: 10.3390/vaccines10091509. Vaccines (Basel). 2022. PMID: 36146586 Free PMC article.
-
T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals.Sci Transl Med. 2021 Apr 21;13(590):eabf7517. doi: 10.1126/scitranslmed.abf7517. Epub 2021 Mar 15. Sci Transl Med. 2021. PMID: 33723016 Free PMC article.
-
Accumulation of mutations in antibody and CD8 T cell epitopes in a B cell depleted lymphoma patient with chronic SARS-CoV-2 infection.Nat Commun. 2022 Sep 23;13(1):5586. doi: 10.1038/s41467-022-32772-5. Nat Commun. 2022. PMID: 36151076 Free PMC article.
-
Longitudinal Analysis and Comparison of Six Serological Assays up to Eight Months Post-COVID-19 Diagnosis.J Clin Med. 2021 Apr 21;10(9):1815. doi: 10.3390/jcm10091815. J Clin Med. 2021. PMID: 33919328 Free PMC article.
References
-
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. - PubMed
-
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. - PubMed
-
- Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci. Immunol. 2020;5:eabd6160. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

