Molecular Mechanisms of DUBs Regulation in Signaling and Disease
- PMID: 33498168
- PMCID: PMC7863924
- DOI: 10.3390/ijms22030986
Molecular Mechanisms of DUBs Regulation in Signaling and Disease
Abstract
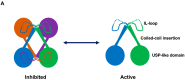
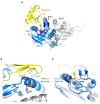
The large family of deubiquitinating enzymes (DUBs) are involved in the regulation of a plethora of processes carried out inside the cell by protein ubiquitination. Ubiquitination is a basic pathway responsible for the correct protein homeostasis in the cell, which could regulate the fate of proteins through the ubiquitin-proteasome system (UPS). In this review we will focus on recent advances on the molecular mechanisms and specificities found for some types of DUBs enzymes, highlighting illustrative examples in which the regulatory mechanism for DUBs has been understood in depth at the molecular level by structural biology. DUB proteases are responsible for cleavage and regulation of the multiple types of ubiquitin linkages that can be synthesized inside the cell, known as the ubiquitin-code, which are tightly connected to specific substrate functions. We will display some strategies carried out by members of different DUB families to provide specificity on the cleavage of particular ubiquitin linkages. Finally, we will also discuss recent progress made for the development of drug compounds targeting DUB proteases, which are usually correlated to the progress of many pathologies such as cancer and neurodegenerative diseases.
Keywords: DUBs; UPS; USPs; deubiquitinating enzymes; proteasome; protein degradation; structural analysis; ubiquitin; ubiquitin-code.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

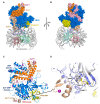






Similar articles
-
Deubiquitinating enzymes (DUBs): Regulation, homeostasis, and oxidative stress response.J Biol Chem. 2021 Sep;297(3):101077. doi: 10.1016/j.jbc.2021.101077. Epub 2021 Aug 12. J Biol Chem. 2021. PMID: 34391779 Free PMC article. Review.
-
DUBs, Hypoxia, and Cancer.Trends Cancer. 2019 Oct;5(10):632-653. doi: 10.1016/j.trecan.2019.08.005. Epub 2019 Oct 11. Trends Cancer. 2019. PMID: 31706510 Review.
-
Breaking the chains: deubiquitylating enzyme specificity begets function.Nat Rev Mol Cell Biol. 2019 Jun;20(6):338-352. doi: 10.1038/s41580-019-0099-1. Nat Rev Mol Cell Biol. 2019. PMID: 30733604 Review.
-
Dissenting degradation: Deubiquitinases in cell cycle and cancer.Semin Cancer Biol. 2020 Dec;67(Pt 2):145-158. doi: 10.1016/j.semcancer.2020.03.008. Epub 2020 Mar 20. Semin Cancer Biol. 2020. PMID: 32201366 Free PMC article. Review.
-
Mechanisms of Deubiquitinase Specificity and Regulation.Annu Rev Biochem. 2017 Jun 20;86:159-192. doi: 10.1146/annurev-biochem-061516-044916. Epub 2017 May 12. Annu Rev Biochem. 2017. PMID: 28498721 Review.
Cited by
-
Deubiquitinase USP1 enhances CCAAT/enhancer-binding protein beta (C/EBPβ) stability and accelerates adipogenesis and lipid accumulation.Cell Death Dis. 2023 Nov 27;14(11):776. doi: 10.1038/s41419-023-06317-7. Cell Death Dis. 2023. PMID: 38012162 Free PMC article.
-
OTULIN Can Improve Spinal Cord Injury by the NF-κB and Wnt/β-Catenin Signaling Pathways.Mol Neurobiol. 2024 Nov;61(11):8820-8830. doi: 10.1007/s12035-024-04134-3. Epub 2024 Apr 2. Mol Neurobiol. 2024. PMID: 38561559 Review.
-
Ubiquitin-specific proteases in inflammatory bowel disease-related signalling pathway regulation.Cell Death Dis. 2022 Feb 10;13(2):139. doi: 10.1038/s41419-022-04566-6. Cell Death Dis. 2022. PMID: 35145062 Free PMC article. Review.
-
An Overview of the Deubiquitinase USP53: A Promising Diagnostic Marker and Therapeutic Target.Curr Protein Pept Sci. 2024;25(9):708-718. doi: 10.2174/0113892037292440240518194922. Curr Protein Pept Sci. 2024. PMID: 39300775 Free PMC article. Review.
-
Deubiquitination complex platform: A plausible mechanism for regulating the substrate specificity of deubiquitinating enzymes.Acta Pharm Sin B. 2023 Jul;13(7):2955-2962. doi: 10.1016/j.apsb.2023.02.019. Epub 2023 Mar 4. Acta Pharm Sin B. 2023. PMID: 37521861 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

