Binding of the SARS-CoV-2 spike protein to glycans
- PMID: 33495714
- PMCID: PMC7816574
- DOI: 10.1016/j.scib.2021.01.010
Binding of the SARS-CoV-2 spike protein to glycans
Abstract
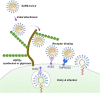
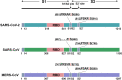
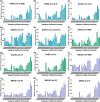
The pandemic of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a high number of deaths in the world. To combat it, it is necessary to develop a better understanding of how the virus infects host cells. Infection normally starts with the attachment of the virus to cell-surface glycans like heparan sulfate (HS) and sialic acid-containing glycolipids/glycoproteins. In this study, we examined and compared the binding of the subunits and spike (S) proteins of SARS-CoV-2, SARS-CoV, and Middle East respiratory disease (MERS)-CoV to these glycans. Our results revealed that the S proteins and subunits can bind to HS in a sulfation-dependent manner and no binding with sialic acid residues was detected. Overall, this work suggests that HS binding may be a general mechanism for the attachment of these coronaviruses to host cells, and supports the potential importance of HS in infection and in the development of antiviral agents against these viruses.
Keywords: Glycan microarray; Heparan sulfate; SARS-CoV-2 S protein; Sialic acid; Sulfation; Surface plasmon resonance.
© 2021 Science China Press. Published by Elsevier B.V. and Science China Press. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

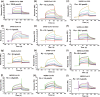
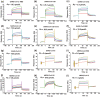
Similar articles
-
Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2.Nat Chem Biol. 2022 Jan;18(1):81-90. doi: 10.1038/s41589-021-00924-1. Epub 2021 Nov 9. Nat Chem Biol. 2022. PMID: 34754101
-
Man-Specific Lectins from Plants, Fungi, Algae and Cyanobacteria, as Potential Blockers for SARS-CoV, MERS-CoV and SARS-CoV-2 (COVID-19) Coronaviruses: Biomedical Perspectives.Cells. 2021 Jun 28;10(7):1619. doi: 10.3390/cells10071619. Cells. 2021. PMID: 34203435 Free PMC article. Review.
-
Heparan Sulfate Facilitates Spike Protein-Mediated SARS-CoV-2 Host Cell Invasion and Contributes to Increased Infection of SARS-CoV-2 G614 Mutant and in Lung Cancer.Front Mol Biosci. 2021 Jun 11;8:649575. doi: 10.3389/fmolb.2021.649575. eCollection 2021. Front Mol Biosci. 2021. PMID: 34179075 Free PMC article.
-
Natural and Recombinant SARS-CoV-2 Isolates Rapidly Evolve In Vitro to Higher Infectivity through More Efficient Binding to Heparan Sulfate and Reduced S1/S2 Cleavage.J Virol. 2021 Oct 13;95(21):e0135721. doi: 10.1128/JVI.01357-21. Epub 2021 Aug 18. J Virol. 2021. PMID: 34406867 Free PMC article.
-
SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction.Int J Mol Sci. 2020 Jun 26;21(12):4549. doi: 10.3390/ijms21124549. Int J Mol Sci. 2020. PMID: 32604730 Free PMC article. Review.
Cited by
-
Single-molecule imaging reveals allosteric stimulation of SARS-CoV-2 spike receptor binding domain by host sialic acid.Sci Adv. 2024 Jul 19;10(29):eadk4920. doi: 10.1126/sciadv.adk4920. Epub 2024 Jul 17. Sci Adv. 2024. PMID: 39018397 Free PMC article.
-
Benzene with Alkyl Chains Is a Universal Scaffold for Multivalent Virucidal Antivirals.ACS Cent Sci. 2024 Apr 4;10(5):1012-1021. doi: 10.1021/acscentsci.4c00054. eCollection 2024 May 22. ACS Cent Sci. 2024. PMID: 38799657 Free PMC article.
-
Back to the Basics of SARS-CoV-2 Biochemistry: Microvascular Occlusive Glycan Bindings Govern Its Morbidities and Inform Therapeutic Responses.Viruses. 2024 Apr 22;16(4):647. doi: 10.3390/v16040647. Viruses. 2024. PMID: 38675987 Free PMC article. Review.
-
Glycosaminoglycan microarrays for studying glycosaminoglycan-protein systems.Carbohydr Polym. 2024 Jul 1;335:122106. doi: 10.1016/j.carbpol.2024.122106. Epub 2024 Mar 29. Carbohydr Polym. 2024. PMID: 38616080 Review.
-
Sulfated Glycans Inhibit the Interaction of MERS-CoV Receptor Binding Domain with Heparin.Viruses. 2024 Feb 2;16(2):237. doi: 10.3390/v16020237. Viruses. 2024. PMID: 38400013 Free PMC article.
References
-
- Drosten C., Günther S., Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek T.G., Erdman D., Goldsmith C.S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. - PubMed
-
- Zaki A.M., van Boheemen S., Bestebroer T.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

