Mechanism of SARS-CoV-2 polymerase stalling by remdesivir
- PMID: 33436624
- PMCID: PMC7804290
- DOI: 10.1038/s41467-020-20542-0
Mechanism of SARS-CoV-2 polymerase stalling by remdesivir
Abstract
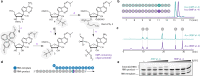
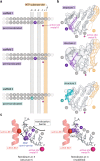
Remdesivir is the only FDA-approved drug for the treatment of COVID-19 patients. The active form of remdesivir acts as a nucleoside analog and inhibits the RNA-dependent RNA polymerase (RdRp) of coronaviruses including SARS-CoV-2. Remdesivir is incorporated by the RdRp into the growing RNA product and allows for addition of three more nucleotides before RNA synthesis stalls. Here we use synthetic RNA chemistry, biochemistry and cryo-electron microscopy to establish the molecular mechanism of remdesivir-induced RdRp stalling. We show that addition of the fourth nucleotide following remdesivir incorporation into the RNA product is impaired by a barrier to further RNA translocation. This translocation barrier causes retention of the RNA 3'-nucleotide in the substrate-binding site of the RdRp and interferes with entry of the next nucleoside triphosphate, thereby stalling RdRp. In the structure of the remdesivir-stalled state, the 3'-nucleotide of the RNA product is matched and located with the template base in the active center, and this may impair proofreading by the viral 3'-exonuclease. These mechanistic insights should facilitate the quest for improved antivirals that target coronavirus replication.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Inhibition of SARS-CoV-2 polymerase by nucleotide analogs from a single-molecule perspective.Elife. 2021 Oct 7;10:e70968. doi: 10.7554/eLife.70968. Elife. 2021. PMID: 34617885 Free PMC article.
-
Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites.Antiviral Res. 2020 Jun;178:104793. doi: 10.1016/j.antiviral.2020.104793. Epub 2020 Apr 10. Antiviral Res. 2020. PMID: 32283108 Free PMC article.
-
Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action.J Biol Chem. 2020 Nov 20;295(47):16156-16165. doi: 10.1074/jbc.AC120.015720. Epub 2020 Sep 23. J Biol Chem. 2020. PMID: 32967965 Free PMC article.
-
RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19.Biochem Biophys Res Commun. 2021 Jan 29;538:47-53. doi: 10.1016/j.bbrc.2020.08.116. Epub 2020 Sep 4. Biochem Biophys Res Commun. 2021. PMID: 32943188 Free PMC article. Review.
-
Remdesivir for the treatment of Covid-19: the value of biochemical studies.Curr Opin Virol. 2021 Aug;49:81-85. doi: 10.1016/j.coviro.2021.04.014. Epub 2021 May 12. Curr Opin Virol. 2021. PMID: 34052732 Free PMC article. Review.
Cited by
-
Characterisation of SARS-CoV-2 genomic variation in response to molnupiravir treatment in the AGILE Phase IIa clinical trial.Nat Commun. 2022 Nov 26;13(1):7284. doi: 10.1038/s41467-022-34839-9. Nat Commun. 2022. PMID: 36435798 Free PMC article. Clinical Trial.
-
Remdesivir inhibits Porcine epidemic diarrhea virus infection in vitro.Heliyon. 2023 Nov 2;9(11):e21468. doi: 10.1016/j.heliyon.2023.e21468. eCollection 2023 Nov. Heliyon. 2023. PMID: 38027806 Free PMC article.
-
Computational designing of a peptide that potentially blocks the entry of SARS-CoV, SARS-CoV-2 and MERS-CoV.PLoS One. 2021 May 18;16(5):e0251913. doi: 10.1371/journal.pone.0251913. eCollection 2021. PLoS One. 2021. PMID: 34003827 Free PMC article.
-
COVID-19 and Gut Injury.Nutrients. 2022 Oct 20;14(20):4409. doi: 10.3390/nu14204409. Nutrients. 2022. PMID: 36297092 Free PMC article. Review.
-
Effectiveness of Remdesivir in Comparison with Five Approved Antiviral Drugs for Inhibition of RdRp in Combat with SARS-CoV-2.Iran J Sci Technol Trans A Sci. 2022;46(5):1359-1367. doi: 10.1007/s40995-022-01364-9. Epub 2022 Sep 24. Iran J Sci Technol Trans A Sci. 2022. PMID: 36187298 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

