Hepatic NOD2 promotes hepatocarcinogenesis via a RIP2-mediated proinflammatory response and a novel nuclear autophagy-mediated DNA damage mechanism
- PMID: 33413510
- PMCID: PMC7791875
- DOI: 10.1186/s13045-020-01028-4
Hepatic NOD2 promotes hepatocarcinogenesis via a RIP2-mediated proinflammatory response and a novel nuclear autophagy-mediated DNA damage mechanism
Abstract
Background: Key hepatic molecules linking gut dysbiosis and hepatocarcinogenesis remain largely unknown. Gut-derived gut microbiota contains pathogen-associated molecular patterns (PAMPs) that may circulate into the liver and, consequently, be recognized by hepatic pattern recognition receptors (PRRs). NOD2, a general intracellular PRR, recognizes muramyl dipeptide (MDP), present in both gram (+) and gram (-) bacteria. Here, we investigated the role of NOD2 as a molecular sensor translating gut dysbiosis signaling into hepatocarcinogenesis.
Methods: NOD2 expression was measured in clinical hepatocellular carcinoma (HCC) samples using qPCR (80 pairs), western blotting (30 pairs) and immunostaining (141 pairs). The role of NOD2 in hepatocarcinogenesis was examined in the hepatocyte-specific Nod2-knockout (Nod2△hep), Rip2-knockout (Rip2△hep), Lamin A/C-knockout (Lamn△hep) and Rip2/Lamin A/C double-knockout (Rip2/Lamn△hep) mice models of diethylnitrosamine (DEN)/CCl4-induced HCC.
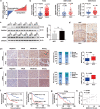
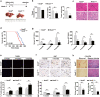
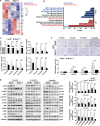
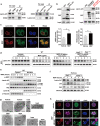
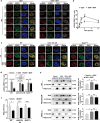
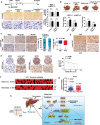
Results: NOD2 was upregulated and activated in HCC samples, and high NOD2 expression correlated with poor prognosis in HCC patients. Hepatic NOD2 deletion in vivo decreased DEN/CCl4-induced HCC by reducing the inflammatory response, DNA damage and genomic instability. NOD2 activation increased liver inflammation via RIP2-dependent activation of the MAPK, NF-κB and STAT3 pathways. Notably, a novel RIP2-independent mechanism was discovered, whereby NOD2 activation induces the nuclear autophagy pathway. We showed that NOD2 undergoes nuclear transport and directly binds to a component of nuclear laminae, lamin A/C, to promote its protein degradation, leading to impaired DNA damage repair and increased genomic instability.
Conclusions: We reveal a novel bridge, bacterial sensor NOD2, linking gut-derived microbial metabolites to hepatocarcinogenesis via induction of the inflammatory response and nuclear autophagy. Thus, we propose hepatic NOD2 as a promising therapeutic target against HCC.
Keywords: C; DNA damage; Hepatocellular carcinoma; Lamin A; NOD2; Nuclear autophagy; RIP2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy.J Biol Chem. 2012 Jul 20;287(30):25565-76. doi: 10.1074/jbc.M111.326835. Epub 2012 Jun 4. J Biol Chem. 2012. PMID: 22665475 Free PMC article.
-
NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2.J Biol Chem. 2007 Dec 14;282(50):36223-9. doi: 10.1074/jbc.M703079200. Epub 2007 Oct 18. J Biol Chem. 2007. PMID: 17947236
-
RIP2 filament formation is required for NOD2 dependent NF-κB signalling.Nat Commun. 2018 Oct 2;9(1):4043. doi: 10.1038/s41467-018-06451-3. Nat Commun. 2018. PMID: 30279485 Free PMC article.
-
Insights into the molecular basis of the NOD2 signalling pathway.Open Biol. 2014 Dec;4(12):140178. doi: 10.1098/rsob.140178. Open Biol. 2014. PMID: 25520185 Free PMC article. Review.
-
RIP2 activity in inflammatory disease and implications for novel therapeutics.J Leukoc Biol. 2013 Nov;94(5):927-32. doi: 10.1189/jlb.0213109. Epub 2013 Jun 21. J Leukoc Biol. 2013. PMID: 23794710 Free PMC article. Review.
Cited by
-
Mechanism of RIP2 enhancing stemness of glioma cells induces temozolomide resistance.CNS Neurosci Ther. 2022 Dec;28(12):2319-2330. doi: 10.1111/cns.13981. Epub 2022 Oct 2. CNS Neurosci Ther. 2022. PMID: 36184801 Free PMC article.
-
Primary and Secondary micro-RNA Modulation the Extrinsic Pathway of Apoptosis in Hepatocellular Carcinoma.Mol Biol. 2023;57(2):165-175. doi: 10.1134/S0026893323020103. Epub 2023 Apr 26. Mol Biol. 2023. PMID: 37128213 Free PMC article.
-
Intratumor microbiome in cancer progression: current developments, challenges and future trends.Biomark Res. 2022 May 31;10(1):37. doi: 10.1186/s40364-022-00381-5. Biomark Res. 2022. PMID: 35642013 Free PMC article. Review.
-
A pyroptosis-related gene signature predicts prognosis and immune microenvironment in hepatocellular carcinoma.World J Surg Oncol. 2022 Jun 3;20(1):179. doi: 10.1186/s12957-022-02617-y. World J Surg Oncol. 2022. PMID: 35659304 Free PMC article.
-
Pattern recognition receptors in health and diseases.Signal Transduct Target Ther. 2021 Aug 4;6(1):291. doi: 10.1038/s41392-021-00687-0. Signal Transduct Target Ther. 2021. PMID: 34344870 Free PMC article. Review.
References
-
- Yu L, Yan H, Liu Q, Yang W, Wu H, Dong W, et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology (Baltimore, Md.). 2010;52:1322–1333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

