Single Mutation in the NFU1 Gene Metabolically Reprograms Pulmonary Artery Smooth Muscle Cells
- PMID: 33297749
- PMCID: PMC7837686
- DOI: 10.1161/ATVBAHA.120.314655
Single Mutation in the NFU1 Gene Metabolically Reprograms Pulmonary Artery Smooth Muscle Cells
Abstract
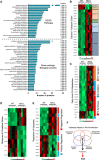
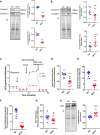
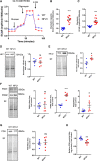
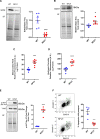
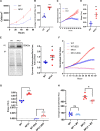
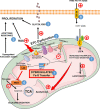
Objective: NFU1 is a mitochondrial iron-sulfur scaffold protein, involved in iron-sulfur assembly and transfer to complex II and LAS (lipoic acid synthase). Patients with the point mutation NFU1G208C and CRISPR/CAS9 (clustered regularly interspaced short palindromic repeats/clustered regularly interspaced short palindromic repeat-associated 9)-generated rats develop mitochondrial dysfunction leading to pulmonary arterial hypertension. However, the mechanistic understanding of pulmonary vascular proliferation due to a single mutation in NFU1 remains unresolved. Approach and Results: Quantitative proteomics of isolated mitochondria showed the entire phenotypic transformation of NFU1G206C rats with a disturbed mitochondrial proteomic landscape, involving significant changes in the expression of 208 mitochondrial proteins. The NFU1 mutation deranged the expression pattern of electron transport proteins, resulting in a significant decrease in mitochondrial respiration. Reduced reliance on mitochondrial respiration amplified glycolysis in pulmonary artery smooth muscle cell (PASMC) and activated GPD (glycerol-3-phosphate dehydrogenase), linking glycolysis to oxidative phosphorylation and lipid metabolism. Decreased PDH (pyruvate dehydrogenase) activity due to the lipoic acid shortage is compensated by increased fatty acid metabolism and oxidation. PASMC became dependent on extracellular fatty acid sources due to upregulated transporters such as CD36 (cluster of differentiation 36) and CPT (carnitine palmitoyltransferase)-1. Finally, the NFU1 mutation produced a dysregulated antioxidant system in the mitochondria, leading to increased reactive oxygen species levels. PASMC from NFU1 rats showed apoptosis resistance, increased anaplerosis, and attained a highly proliferative phenotype. Attenuation of mitochondrial reactive oxygen species by mitochondrial-targeted antioxidant significantly decreased PASMC proliferation.
Conclusions: The alteration in iron-sulfur metabolism completely transforms the proteomic landscape of the mitochondria, leading toward metabolic plasticity and redistribution of energy sources to the acquisition of a proliferative phenotype by the PASMC.
Keywords: glycolysis; metabolism; mitochondria; myocytes; pulmonary artery; smooth muscle.
Conflict of interest statement
None.
Figures


Similar articles
-
Rats with a Human Mutation of NFU1 Develop Pulmonary Hypertension.Am J Respir Cell Mol Biol. 2020 Feb;62(2):231-242. doi: 10.1165/rcmb.2019-0065OC. Am J Respir Cell Mol Biol. 2020. PMID: 31461310 Free PMC article.
-
Reoxygenation Reverses Hypoxic Pulmonary Arterial Remodeling by Inducing Smooth Muscle Cell Apoptosis via Reactive Oxygen Species-Mediated Mitochondrial Dysfunction.J Am Heart Assoc. 2017 Jun 23;6(6):e005602. doi: 10.1161/JAHA.117.005602. J Am Heart Assoc. 2017. PMID: 28645933 Free PMC article.
-
CPT1 regulates the proliferation of pulmonary artery smooth muscle cells through the AMPK-p53-p21 pathway in pulmonary arterial hypertension.Mol Cell Biochem. 2019 May;455(1-2):169-183. doi: 10.1007/s11010-018-3480-z. Epub 2018 Dec 3. Mol Cell Biochem. 2019. PMID: 30511343
-
Dynamic and diverse changes in the functional properties of vascular smooth muscle cells in pulmonary hypertension.Cardiovasc Res. 2018 Mar 15;114(4):551-564. doi: 10.1093/cvr/cvy004. Cardiovasc Res. 2018. PMID: 29385432 Free PMC article. Review.
-
The metabolic theory of pulmonary arterial hypertension.Circ Res. 2014 Jun 20;115(1):148-64. doi: 10.1161/CIRCRESAHA.115.301130. Circ Res. 2014. PMID: 24951764 Review.
Cited by
-
Mitochondria as a primary determinant of angiogenic modality in pulmonary arterial hypertension.J Exp Med. 2024 Nov 4;221(11):e20231568. doi: 10.1084/jem.20231568. Epub 2024 Sep 25. J Exp Med. 2024. PMID: 39320470
-
Patient-specific variants of NFU1/NFU-1 disrupt cholinergic signaling in a model of multiple mitochondrial dysfunctions syndrome 1.Dis Model Mech. 2023 Feb 1;16(2):dmm049594. doi: 10.1242/dmm.049594. Epub 2023 Feb 1. Dis Model Mech. 2023. PMID: 36645076 Free PMC article.
-
Metabolism, Mitochondrial Dysfunction, and Redox Homeostasis in Pulmonary Hypertension.Antioxidants (Basel). 2022 Feb 21;11(2):428. doi: 10.3390/antiox11020428. Antioxidants (Basel). 2022. PMID: 35204311 Free PMC article. Review.
-
Proteomic Signatures of Diffuse and Intestinal Subtypes of Gastric Cancer.Cancers (Basel). 2021 Nov 25;13(23):5930. doi: 10.3390/cancers13235930. Cancers (Basel). 2021. PMID: 34885041 Free PMC article.
-
Study on the Regulatory Mechanism of the PDK1-Mediated TGF-β/Smad Signaling Pathway in Hypoxia-Induced Yak Lungs.Animals (Basel). 2024 Aug 21;14(16):2422. doi: 10.3390/ani14162422. Animals (Basel). 2024. PMID: 39199957 Free PMC article.
References
-
- Stenmark KR, Fagan KA, Frid MG. Hypoxia-induced pulmonary vascular remodeling: cellular and molecular mechanisms. Circ Res. 2006;99:675–691. doi: 10.1161/01.RES.0000243584.45145.3f - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

