Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis
- PMID: 33217318
- PMCID: PMC7816607
- DOI: 10.1016/j.molcel.2020.10.032
Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis
Abstract
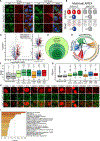
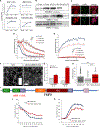
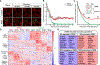
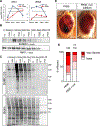
Stress granules (SGs) are cytoplasmic assemblies of proteins and non-translating mRNAs. Whereas much has been learned about SG formation, a major gap remains in understanding the compositional changes SGs undergo during normal disassembly and under disease conditions. Here, we address this gap by proteomic dissection of the SG temporal disassembly sequence using multi-bait APEX proximity proteomics. We discover 109 novel SG proteins and characterize distinct SG substructures. We reveal dozens of disassembly-engaged proteins (DEPs), some of which play functional roles in SG disassembly, including small ubiquitin-like modifier (SUMO) conjugating enzymes. We further demonstrate that SUMOylation regulates SG disassembly and SG formation. Parallel proteomics with amyotrophic lateral sclerosis (ALS)-associated C9ORF72 dipeptides uncovered attenuated DEP recruitment during SG disassembly and impaired SUMOylation. Accordingly, SUMO activity ameliorated C9ORF72-ALS-related neurodegeneration in Drosophila. By dissecting the SG spatiotemporal proteomic landscape, we provide an in-depth resource for future work on SG function and reveal basic and disease-relevant mechanisms of SG disassembly.
Keywords: ALS; APEX; RNA binding proteins; amyotrophic lateral sclerosis; condensates; membraneless organelles; neurodegeneration; phase separation; stress granules; sumoylation.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


Similar articles
-
Stress Granule Homeostasis, Aberrant Phase Transition, and Amyotrophic Lateral Sclerosis.ACS Chem Neurosci. 2022 Aug 17;13(16):2356-2370. doi: 10.1021/acschemneuro.2c00262. Epub 2022 Jul 29. ACS Chem Neurosci. 2022. PMID: 35905138 Review.
-
Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis.Nat Med. 2018 Aug;24(8):1136-1142. doi: 10.1038/s41591-018-0071-1. Epub 2018 Jun 25. Nat Med. 2018. PMID: 29942091 Free PMC article.
-
Traumatic injury induces stress granule formation and enhances motor dysfunctions in ALS/FTD models.Hum Mol Genet. 2018 Apr 15;27(8):1366-1381. doi: 10.1093/hmg/ddy047. Hum Mol Genet. 2018. PMID: 29432563 Free PMC article.
-
Conserved metabolite regulation of stress granule assembly via AdoMet.J Cell Biol. 2020 Aug 3;219(8):e201904141. doi: 10.1083/jcb.201904141. J Cell Biol. 2020. PMID: 32609300 Free PMC article.
-
Drosophila models of amyotrophic lateral sclerosis with defects in RNA metabolism.Brain Res. 2018 Aug 15;1693(Pt A):109-120. doi: 10.1016/j.brainres.2018.04.043. Epub 2018 May 9. Brain Res. 2018. PMID: 29752901 Review.
Cited by
-
APEX Proximity Labeling of Stress Granule Proteins.Methods Mol Biol. 2022;2428:381-399. doi: 10.1007/978-1-0716-1975-9_23. Methods Mol Biol. 2022. PMID: 35171492 Free PMC article.
-
Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients.Antioxidants (Basel). 2022 Jun 8;11(6):1129. doi: 10.3390/antiox11061129. Antioxidants (Basel). 2022. PMID: 35740026 Free PMC article.
-
K-Homology Type Splicing Regulatory Protein: Mechanism of Action in Cancer and Immune Disorders.Crit Rev Eukaryot Gene Expr. 2024;34(1):75-87. doi: 10.1615/CritRevEukaryotGeneExpr.2023048085. Crit Rev Eukaryot Gene Expr. 2024. PMID: 37824394 Free PMC article.
-
Stressful steps: Progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules.Mol Cell. 2022 Jul 21;82(14):2544-2556. doi: 10.1016/j.molcel.2022.05.014. Epub 2022 Jun 3. Mol Cell. 2022. PMID: 35662398 Free PMC article. Review.
-
Novel Components of the Stress Assembly Sec Body Identified by Proximity Labeling.Cells. 2023 Mar 30;12(7):1055. doi: 10.3390/cells12071055. Cells. 2023. PMID: 37048128 Free PMC article.
References
-
- Alberti S, and Dormann D (2019). Liquid-liquid phase separation in disease. Annu. Rev. Genet 53, 171–194. - PubMed
-
- Anderson P, and Kedersha N (2008). Stress granules: the Tao of RNA triage. Trends Biochem. Sci 33, 141–150. - PubMed
-
- Babu MM, Kriwacki RW, and Pappu RV (2012). Structural biology. Versatility from protein disorder. Science 337, 1460–1461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

