Commensal Microbiota Modulation of Natural Resistance to Virus Infection
- PMID: 33212011
- PMCID: PMC7799371
- DOI: 10.1016/j.cell.2020.10.047
Commensal Microbiota Modulation of Natural Resistance to Virus Infection
Abstract
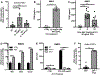
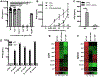
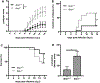
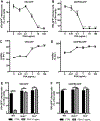
Interferon (IFN)-Is are crucial mediators of antiviral immunity and homeostatic immune system regulation. However, the source of IFN-I signaling under homeostatic conditions is unclear. We discovered that commensal microbes regulate the IFN-I response through induction of IFN-β by colonic DCs. Moreover, the mechanism by which a specific commensal microbe induces IFN-β was identified. Outer membrane (OM)-associated glycolipids of gut commensal microbes belonging to the Bacteroidetes phylum induce expression of IFN-β. Using Bacteroides fragilis and its OM-associated polysaccharide A, we determined that IFN-β expression was induced via TLR4-TRIF signaling. Antiviral activity of this purified microbial molecule against infection with either vesicular stomatitis virus (VSV) or influenza was demonstrated to be dependent on the induction of IFN-β. In a murine VSV infection model, commensal-induced IFN-β regulated natural resistance to virus infection. Due to the physiological importance of IFN-Is, discovery of an IFN-β-inducing microbial molecule represents a potential approach for the treatment of some human diseases.
Keywords: Bacteroides; dendritic cell; immune modulation; microbiome; type I interferon; virus infection.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests Two patent applications have been filed by Harvard University with relation to this work. D.L.K. and K.L.S. are listed as inventors on both.
Figures



Similar articles
-
MAVS-dependent IRF3/7 bypass of interferon β-induction restricts the response to measles infection in CD150Tg mouse bone marrow-derived dendritic cells.Mol Immunol. 2014 Feb;57(2):100-10. doi: 10.1016/j.molimm.2013.08.007. Epub 2013 Oct 4. Mol Immunol. 2014. PMID: 24096085
-
Concomitant TLR/RLH signaling of radioresistant and radiosensitive cells is essential for protection against vesicular stomatitis virus infection.J Immunol. 2014 Sep 15;193(6):3045-54. doi: 10.4049/jimmunol.1400959. Epub 2014 Aug 15. J Immunol. 2014. PMID: 25127863
-
Impairment of the retinoic acid-inducible gene-I-IFN-β signaling pathway in chronic hepatitis B virus infection.Int J Mol Med. 2012 Dec;30(6):1498-504. doi: 10.3892/ijmm.2012.1131. Epub 2012 Sep 19. Int J Mol Med. 2012. PMID: 22992810
-
The Intestinal Commensal, Bacteroides fragilis, Modulates Host Responses to Viral Infection and Therapy: Lessons for Exploration during Mycobacterium tuberculosis Infection.Infect Immun. 2022 Jan 25;90(1):e0032121. doi: 10.1128/IAI.00321-21. Epub 2021 Oct 4. Infect Immun. 2022. PMID: 34606367 Free PMC article. Review.
-
Microbiota regulation of viral infections through interferon signaling.Trends Microbiol. 2022 Aug;30(8):778-792. doi: 10.1016/j.tim.2022.01.007. Epub 2022 Feb 6. Trends Microbiol. 2022. PMID: 35135717 Free PMC article. Review.
Cited by
-
Microbiome Dynamics: A Paradigm Shift in Combatting Infectious Diseases.J Pers Med. 2024 Feb 18;14(2):217. doi: 10.3390/jpm14020217. J Pers Med. 2024. PMID: 38392650 Free PMC article. Review.
-
Recent advances on smart glycoconjugate vaccines in infections and cancer.FEBS J. 2022 Jul;289(14):4251-4303. doi: 10.1111/febs.15909. Epub 2021 Jun 1. FEBS J. 2022. PMID: 33934527 Free PMC article. Review.
-
RBM14 inhibits the replication of porcine epidemic diarrhea virus by recruiting p62 to degrade nucleocapsid protein through the activation of autophagy and interferon pathway.J Virol. 2024 Mar 19;98(3):e0018224. doi: 10.1128/jvi.00182-24. Epub 2024 Feb 27. J Virol. 2024. PMID: 38411947 Free PMC article.
-
N protein of PEDV plays chess game with host proteins by selective autophagy.Autophagy. 2023 Aug;19(8):2338-2352. doi: 10.1080/15548627.2023.2181615. Epub 2023 Mar 2. Autophagy. 2023. PMID: 36861818 Free PMC article.
-
Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors.Cell. 2022 Mar 31;185(7):1157-1171.e22. doi: 10.1016/j.cell.2022.02.002. Epub 2022 Mar 7. Cell. 2022. PMID: 35259335 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

