Clover: a clustering-oriented de novo assembler for Illumina sequences
- PMID: 33203354
- PMCID: PMC7672897
- DOI: 10.1186/s12859-020-03788-9
Clover: a clustering-oriented de novo assembler for Illumina sequences
Abstract
Background: Next-generation sequencing technologies revolutionized genomics by producing high-throughput reads at low cost, and this progress has prompted the recent development of de novo assemblers. Multiple assembly methods based on de Bruijn graph have been shown to be efficient for Illumina reads. However, the sequencing errors generated by the sequencer complicate analysis of de novo assembly and influence the quality of downstream genomic researches.
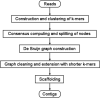
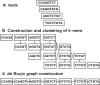
Results: In this paper, we develop a de Bruijn assembler, called Clover (clustering-oriented de novo assembler), that utilizes a novel k-mer clustering approach from the overlap-layout-consensus concept to deal with the sequencing errors generated by the Illumina platform. We further evaluate Clover's performance against several de Bruijn graph assemblers (ABySS, SOAPdenovo, SPAdes and Velvet), overlap-layout-consensus assemblers (Bambus2, CABOG and MSR-CA) and string graph assembler (SGA) on three datasets (Staphylococcus aureus, Rhodobacter sphaeroides and human chromosome 14). The results show that Clover achieves a superior assembly quality in terms of corrected N50 and E-size while remaining a significantly competitive in run time except SOAPdenovo. In addition, Clover was involved in the sequencing projects of bacterial genomes Acinetobacter baumannii TYTH-1 and Morganella morganii KT.
Conclusions: The marvel clustering-based approach of Clover that integrates the flexibility of the overlap-layout-consensus approach and the efficiency of the de Bruijn graph method has high potential on de novo assembly. Now, Clover is freely available as open source software from https://oz.nthu.edu.tw/~d9562563/src.html .
Keywords: DNA sequencing; De bruijn graph; De novo genome assembly.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Benchmarking of de novo assembly algorithms for Nanopore data reveals optimal performance of OLC approaches.BMC Genomics. 2016 Aug 22;17 Suppl 7(Suppl 7):507. doi: 10.1186/s12864-016-2895-8. BMC Genomics. 2016. PMID: 27556636 Free PMC article.
-
Evaluating de Bruijn graph assemblers on 454 transcriptomic data.PLoS One. 2012;7(12):e51188. doi: 10.1371/journal.pone.0051188. Epub 2012 Dec 7. PLoS One. 2012. PMID: 23236450 Free PMC article.
-
BASE: a practical de novo assembler for large genomes using long NGS reads.BMC Genomics. 2016 Aug 31;17 Suppl 5(Suppl 5):499. doi: 10.1186/s12864-016-2829-5. BMC Genomics. 2016. PMID: 27586129 Free PMC article.
-
Assembly algorithms for next-generation sequencing data.Genomics. 2010 Jun;95(6):315-27. doi: 10.1016/j.ygeno.2010.03.001. Epub 2010 Mar 6. Genomics. 2010. PMID: 20211242 Free PMC article. Review.
-
The present and future of de novo whole-genome assembly.Brief Bioinform. 2018 Jan 1;19(1):23-40. doi: 10.1093/bib/bbw096. Brief Bioinform. 2018. PMID: 27742661 Review.
Cited by
-
Long noncoding RNA study: Genome-wide approaches.Genes Dis. 2022 Nov 29;10(6):2491-2510. doi: 10.1016/j.gendis.2022.10.024. eCollection 2023 Nov. Genes Dis. 2022. PMID: 37554208 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous