The ER cholesterol sensor SCAP promotes CARTS biogenesis at ER-Golgi membrane contact sites
- PMID: 33156328
- PMCID: PMC7654440
- DOI: 10.1083/jcb.202002150
The ER cholesterol sensor SCAP promotes CARTS biogenesis at ER-Golgi membrane contact sites
Abstract
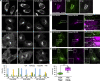
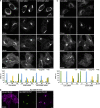
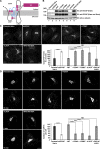
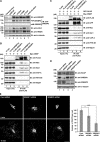
In response to cholesterol deprivation, SCAP escorts SREBP transcription factors from the endoplasmic reticulum to the Golgi complex for their proteolytic activation, leading to gene expression for cholesterol synthesis and uptake. Here, we show that in cholesterol-fed cells, ER-localized SCAP interacts through Sac1 phosphatidylinositol 4-phosphate (PI4P) phosphatase with a VAP-OSBP complex, which mediates counter-transport of ER cholesterol and Golgi PI4P at ER-Golgi membrane contact sites (MCSs). SCAP knockdown inhibited the turnover of PI4P, perhaps due to a cholesterol transport defect, and altered the subcellular distribution of the VAP-OSBP complex. As in the case of perturbation of lipid transfer complexes at ER-Golgi MCSs, SCAP knockdown inhibited the biogenesis of the trans-Golgi network-derived transport carriers CARTS, which was reversed by expression of wild-type SCAP or a Golgi transport-defective mutant, but not of cholesterol sensing-defective mutants. Altogether, our findings reveal a new role for SCAP under cholesterol-fed conditions in the facilitation of CARTS biogenesis via ER-Golgi MCSs, depending on the ER cholesterol.
© 2020 Wakana et al.
Figures
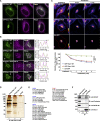


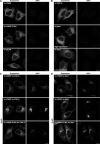
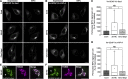
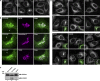
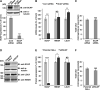
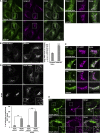
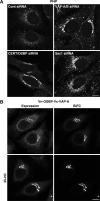
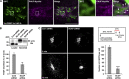
Similar articles
-
Assaying Sterol-Regulated ER-to-Golgi Transport of SREBP Cleavage-Activating Protein Using Immunofluorescence Microscopy.Methods Mol Biol. 2023;2557:755-764. doi: 10.1007/978-1-0716-2639-9_45. Methods Mol Biol. 2023. PMID: 36512249 Free PMC article.
-
Fatostatin blocks ER exit of SCAP but inhibits cell growth in a SCAP-independent manner.J Lipid Res. 2016 Aug;57(8):1564-73. doi: 10.1194/jlr.M069583. Epub 2016 Jun 20. J Lipid Res. 2016. PMID: 27324795 Free PMC article.
-
PAQR3 modulates cholesterol homeostasis by anchoring Scap/SREBP complex to the Golgi apparatus.Nat Commun. 2015 Aug 27;6:8100. doi: 10.1038/ncomms9100. Nat Commun. 2015. PMID: 26311497 Free PMC article.
-
The counterflow transport of sterols and PI4P.Biochim Biophys Acta. 2016 Aug;1861(8 Pt B):940-951. doi: 10.1016/j.bbalip.2016.02.024. Epub 2016 Feb 27. Biochim Biophys Acta. 2016. PMID: 26928592 Review.
-
[Disorder of cholesterol metabolism: regulation of intracellular cholesterol and membrane trafficking].Rinsho Byori. 2010 Dec;58(12):1203-10. Rinsho Byori. 2010. PMID: 21348240 Review. Japanese.
Cited by
-
Calcium flow at ER-TGN contact sites facilitates secretory cargo export.Mol Biol Cell. 2024 Apr 1;35(4):ar50. doi: 10.1091/mbc.E23-03-0099. Epub 2024 Jan 31. Mol Biol Cell. 2024. PMID: 38294859 Free PMC article.
-
Sending out molecules from the TGN.Curr Opin Cell Biol. 2021 Aug;71:55-62. doi: 10.1016/j.ceb.2021.02.005. Epub 2021 Mar 8. Curr Opin Cell Biol. 2021. PMID: 33706234 Free PMC article. Review.
-
NPC1 regulates the distribution of phosphatidylinositol 4-kinases at Golgi and lysosomal membranes.EMBO J. 2021 Jul 1;40(13):e105990. doi: 10.15252/embj.2020105990. Epub 2021 May 21. EMBO J. 2021. PMID: 34019311 Free PMC article.
-
Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets.Viruses. 2023 Apr 29;15(5):1098. doi: 10.3390/v15051098. Viruses. 2023. PMID: 37243184 Free PMC article.
-
Sorting of secretory proteins at the trans-Golgi network by human TGN46.Elife. 2024 Mar 11;12:RP91708. doi: 10.7554/eLife.91708. Elife. 2024. PMID: 38466628 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

