Structure, lipid scrambling activity and role in autophagosome formation of ATG9A
- PMID: 33106659
- PMCID: PMC7718406
- DOI: 10.1038/s41594-020-00520-2
Structure, lipid scrambling activity and role in autophagosome formation of ATG9A
Abstract
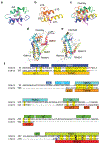
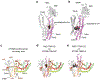
De novo formation of the double-membrane compartment autophagosome is seeded by small vesicles carrying membrane protein autophagy-related 9 (ATG9), the function of which remains unknown. Here we find that ATG9A scrambles phospholipids of membranes in vitro. Cryo-EM structures of human ATG9A reveal a trimer with a solvated central pore, which is connected laterally to the cytosol through the cavity within each protomer. Similarities to ABC exporters suggest that ATG9A could be a transporter that uses the central pore to function. Moreover, molecular dynamics simulation suggests that the central pore opens laterally to accommodate lipid headgroups, thereby enabling lipids to flip. Mutations in the pore reduce scrambling activity and yield markedly smaller autophagosomes, indicating that lipid scrambling by ATG9A is essential for membrane expansion. We propose ATG9A acts as a membrane-embedded funnel to facilitate lipid flipping and to redistribute lipids added to the outer leaflet of ATG9 vesicles, thereby enabling growth into autophagosomes.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures


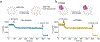
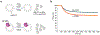



Similar articles
-
Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion.Nat Struct Mol Biol. 2020 Dec;27(12):1185-1193. doi: 10.1038/s41594-020-00518-w. Epub 2020 Oct 26. Nat Struct Mol Biol. 2020. PMID: 33106658
-
Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.Cell Rep. 2020 Jun 30;31(13):107837. doi: 10.1016/j.celrep.2020.107837. Cell Rep. 2020. PMID: 32610138 Free PMC article.
-
Structural basis for lipid transfer by the ATG2A-ATG9A complex.Nat Struct Mol Biol. 2025 Jan;32(1):35-47. doi: 10.1038/s41594-024-01376-6. Epub 2024 Aug 22. Nat Struct Mol Biol. 2025. PMID: 39174844
-
Mammalian autophagy and the plasma membrane.FEBS J. 2017 Mar;284(5):672-679. doi: 10.1111/febs.13931. Epub 2016 Nov 6. FEBS J. 2017. PMID: 27758042 Review.
-
The role of AP-4 in cargo export from the trans-Golgi network and hereditary spastic paraplegia.Biochem Soc Trans. 2020 Oct 30;48(5):1877-1888. doi: 10.1042/BST20190664. Biochem Soc Trans. 2020. PMID: 33084855 Review.
Cited by
-
Morphology of Phagophore Precursors by Correlative Light-Electron Microscopy.Cells. 2022 Sep 30;11(19):3080. doi: 10.3390/cells11193080. Cells. 2022. PMID: 36231043 Free PMC article.
-
Reconstitution of cargo-induced LC3 lipidation in mammalian selective autophagy.Sci Adv. 2021 Apr 23;7(17):eabg4922. doi: 10.1126/sciadv.abg4922. Print 2021 Apr. Sci Adv. 2021. PMID: 33893090 Free PMC article.
-
miRNA family miR-29 inhibits PINK1-PRKN dependent mitophagy via ATG9A.bioRxiv [Preprint]. 2024 Jan 19:2024.01.17.576122. doi: 10.1101/2024.01.17.576122. bioRxiv. 2024. PMID: 38293184 Free PMC article. Preprint.
-
Fission Yeast Autophagy Machinery.Cells. 2022 Mar 24;11(7):1086. doi: 10.3390/cells11071086. Cells. 2022. PMID: 35406650 Free PMC article. Review.
-
Systematic Assessment of the Accuracy of Subunit Counting in Biomolecular Complexes Using Automated Single-Molecule Brightness Analysis.J Phys Chem Lett. 2022 Jan 27;13(3):822-829. doi: 10.1021/acs.jpclett.1c03835. Epub 2022 Jan 19. J Phys Chem Lett. 2022. PMID: 35044771 Free PMC article.
References
Methods-only References
-
- Suloway C et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

