Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity
- PMID: 33082293
- PMCID: PMC7857391
- DOI: 10.1126/science.abd2985
Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity
Abstract
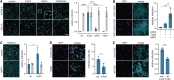
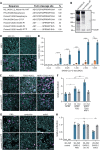
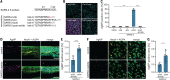
The causative agent of coronavirus disease 2019 (COVID-19) is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). For many viruses, tissue tropism is determined by the availability of virus receptors and entry cofactors on the surface of host cells. In this study, we found that neuropilin-1 (NRP1), known to bind furin-cleaved substrates, significantly potentiates SARS-CoV-2 infectivity, an effect blocked by a monoclonal blocking antibody against NRP1. A SARS-CoV-2 mutant with an altered furin cleavage site did not depend on NRP1 for infectivity. Pathological analysis of olfactory epithelium obtained from human COVID-19 autopsies revealed that SARS-CoV-2 infected NRP1-positive cells facing the nasal cavity. Our data provide insight into SARS-CoV-2 cell infectivity and define a potential target for antiviral intervention.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

Comment in
-
Not only ACE2-the quest for additional host cell mediators of SARS-CoV-2 infection: Neuropilin-1 (NRP1) as a novel SARS-CoV-2 host cell entry mediator implicated in COVID-19.Signal Transduct Target Ther. 2021 Jan 18;6(1):21. doi: 10.1038/s41392-020-00460-9. Signal Transduct Target Ther. 2021. PMID: 33462185 Free PMC article. No abstract available.
Similar articles
-
Neuropilin-1 is a host factor for SARS-CoV-2 infection.Science. 2020 Nov 13;370(6518):861-865. doi: 10.1126/science.abd3072. Epub 2020 Oct 20. Science. 2020. PMID: 33082294 Free PMC article.
-
Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein.J Virol. 2020 Oct 14;94(21):e01062-20. doi: 10.1128/JVI.01062-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32788194 Free PMC article.
-
SARS-CoV-2 host tropism: An in silico analysis of the main cellular factors.Virus Res. 2020 Nov;289:198154. doi: 10.1016/j.virusres.2020.198154. Epub 2020 Sep 9. Virus Res. 2020. PMID: 32918944 Free PMC article.
-
Host Cell and SARS-CoV-2-Associated Molecular Structures and Factors as Potential Therapeutic Targets.Cells. 2021 Sep 15;10(9):2427. doi: 10.3390/cells10092427. Cells. 2021. PMID: 34572076 Free PMC article. Review.
-
COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2.PLoS Pathog. 2020 Aug 21;16(8):e1008762. doi: 10.1371/journal.ppat.1008762. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32822426 Free PMC article. Review.
Cited by
-
N-alkylation of amines for the synthesis of potential antiviral agents: A structural modification approach.Heliyon. 2024 Sep 27;10(19):e38587. doi: 10.1016/j.heliyon.2024.e38587. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39397970 Free PMC article.
-
Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.Nat Commun. 2024 Oct 7;15(1):8574. doi: 10.1038/s41467-024-52808-2. Nat Commun. 2024. PMID: 39375326 Free PMC article.
-
Diesel exhaust particle inhalation in conjunction with high-fat diet consumption alters the expression of pulmonary SARS-COV-2 infection pathways, which is mitigated by probiotic treatment in C57BL/6 male mice.Part Fibre Toxicol. 2024 Sep 29;21(1):40. doi: 10.1186/s12989-024-00601-w. Part Fibre Toxicol. 2024. PMID: 39343929 Free PMC article.
-
Interactions of SARS-CoV-2 with Human Target Cells-A Metabolic View.Int J Mol Sci. 2024 Sep 16;25(18):9977. doi: 10.3390/ijms25189977. Int J Mol Sci. 2024. PMID: 39337465 Free PMC article. Review.
-
Aggravating mechanisms from COVID-19.Virol J. 2024 Sep 27;21(1):228. doi: 10.1186/s12985-024-02506-8. Virol J. 2024. PMID: 39334442 Free PMC article. Review.
References
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating and Research Team , A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 - DOI - PMC - PubMed
-
- Wölfel R., Corman V. M., Guggemos W., Seilmaier M., Zange S., Müller M. A., Niemeyer D., Jones T. C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C., Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). 10.1038/s41586-020-2196-x - DOI - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 - DOI - PMC - PubMed
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e8 (2020). 10.1016/j.cell.2020.02.052 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

