Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor
- PMID: 33077916
- PMCID: PMC9377523
- DOI: 10.1038/s41588-020-00731-9
Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor
Abstract
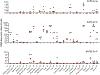
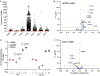
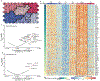
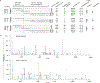
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes COVID-19, utilizes angiotensin-converting enzyme 2 (ACE2) for entry into target cells. ACE2 has been proposed as an interferon-stimulated gene (ISG). Thus, interferon-induced variability in ACE2 expression levels could be important for susceptibility to COVID-19 or its outcomes. Here, we report the discovery of a novel, transcriptionally independent truncated isoform of ACE2, which we designate as deltaACE2 (dACE2). We demonstrate that dACE2, but not ACE2, is an ISG. In The Cancer Genome Atlas, the expression of dACE2 was enriched in squamous tumors of the respiratory, gastrointestinal and urogenital tracts. In vitro, dACE2, which lacks 356 amino-terminal amino acids, was non-functional in binding the SARS-CoV-2 spike protein and as a carboxypeptidase. Our results suggest that the ISG-type induction of dACE2 in IFN-high conditions created by treatments, an inflammatory tumor microenvironment or viral co-infections is unlikely to increase the cellular entry of SARS-CoV-2 and promote infection.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures










Update of
-
Interferons and viruses induce a novel primate-specific isoform dACE2 and not the SARS-CoV-2 receptor ACE2.bioRxiv [Preprint]. 2020 Jul 20:2020.07.19.210955. doi: 10.1101/2020.07.19.210955. bioRxiv. 2020. Update in: Nat Genet. 2020 Dec;52(12):1283-1293. doi: 10.1038/s41588-020-00731-9. PMID: 32743577 Free PMC article. Updated. Preprint.
Similar articles
-
Interferons and viruses induce a novel primate-specific isoform dACE2 and not the SARS-CoV-2 receptor ACE2.bioRxiv [Preprint]. 2020 Jul 20:2020.07.19.210955. doi: 10.1101/2020.07.19.210955. bioRxiv. 2020. Update in: Nat Genet. 2020 Dec;52(12):1283-1293. doi: 10.1038/s41588-020-00731-9. PMID: 32743577 Free PMC article. Updated. Preprint.
-
Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option.Nat Genet. 2020 Dec;52(12):1294-1302. doi: 10.1038/s41588-020-00732-8. Epub 2020 Oct 19. Nat Genet. 2020. PMID: 33077915 Free PMC article.
-
Regulation of IFNα-induced expression of the short ACE2 isoform by ULK1.Mol Immunol. 2022 Jul;147:1-9. doi: 10.1016/j.molimm.2022.04.008. Epub 2022 Apr 27. Mol Immunol. 2022. PMID: 35489289 Free PMC article.
-
Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19.Front Immunol. 2021 Dec 22;12:732690. doi: 10.3389/fimmu.2021.732690. eCollection 2021. Front Immunol. 2021. PMID: 35003058 Free PMC article. Review.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
Cited by
-
A genome-wide arrayed CRISPR screen identifies PLSCR1 as an intrinsic barrier to SARS-CoV-2 entry that recent virus variants have evolved to resist.PLoS Biol. 2024 Sep 24;22(9):e3002767. doi: 10.1371/journal.pbio.3002767. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39316623 Free PMC article.
-
Circulating miRNAs in the Plasma of Post-COVID-19 Patients with Typical Recovery and Those with Long-COVID Symptoms: Regulation of Immune Response-Associated Pathways.Noncoding RNA. 2024 Sep 2;10(5):48. doi: 10.3390/ncrna10050048. Noncoding RNA. 2024. PMID: 39311385 Free PMC article.
-
Human ACE2 Gene Replacement Mice Support SARS-CoV-2 Viral Replication and Nonlethal Disease Progression.Immunohorizons. 2024 Sep 1;8(9):712-720. doi: 10.4049/immunohorizons.2400030. Immunohorizons. 2024. PMID: 39287601 Free PMC article.
-
Impact of the SARS-CoV-2 Spike Protein on the Innate Immune System: A Review.Cureus. 2024 Mar 26;16(3):e57008. doi: 10.7759/cureus.57008. eCollection 2024 Mar. Cureus. 2024. PMID: 38549864 Free PMC article. Review.
-
Soluble ACE2 correlates with severe COVID-19 and can impair antibody responses.iScience. 2024 Feb 24;27(3):109330. doi: 10.1016/j.isci.2024.109330. eCollection 2024 Mar 15. iScience. 2024. PMID: 38496296 Free PMC article.
References
-
- Chua RL et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ZIA CP010201/ImNIH/Intramural NIH HHS/United States
- R21 AG064479/AG/NIA NIH HHS/United States
- 272983813/Deutsche Forschungsgemeinschaft (German Research Foundation)/International
- 416072091/Deutsche Forschungsgemeinschaft (German Research Foundation)/International
- Z99 CA999999/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

