Unexpected specificity within dynamic transcriptional protein-protein complexes
- PMID: 33077600
- PMCID: PMC7959569
- DOI: 10.1073/pnas.2013244117
Unexpected specificity within dynamic transcriptional protein-protein complexes
Abstract
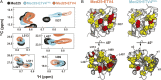
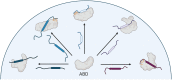
A key functional event in eukaryotic gene activation is the formation of dynamic protein-protein interaction networks between transcriptional activators and transcriptional coactivators. Seemingly incongruent with the tight regulation of transcription, many biochemical and biophysical studies suggest that activators use nonspecific hydrophobic and/or electrostatic interactions to bind to coactivators, with few if any specific contacts. Here a mechanistic dissection of a set of representative dynamic activator•coactivator complexes, comprised of the ETV/PEA3 family of activators and the coactivator Med25, reveals a different molecular recognition model. The data demonstrate that small sequence variations within an activator family significantly redistribute the conformational ensemble of the complex while not affecting overall affinity, and distal residues within the activator-not often considered as contributing to binding-play a key role in mediating conformational redistribution. The ETV/PEA3•Med25 ensembles are directed by specific contacts between the disordered activator and the Med25 interface, which is facilitated by structural shifts of the coactivator binding surface. Taken together, these data highlight the critical role coactivator plasticity plays in recognition of disordered activators and indicate that molecular recognition models of disordered proteins must consider the ability of the binding partners to mediate specificity.
Keywords: ETV/PEA3; Med25; coactivator; protein–protein interactions; transcriptional activator.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
The Mediator complex subunit MED25 is targeted by the N-terminal transactivation domain of the PEA3 group members.Nucleic Acids Res. 2013 May;41(9):4847-59. doi: 10.1093/nar/gkt199. Epub 2013 Mar 26. Nucleic Acids Res. 2013. PMID: 23531547 Free PMC article.
-
Multivalent and Bidirectional Binding of Transcriptional Transactivation Domains to the MED25 Coactivator.Biomolecules. 2020 Aug 19;10(9):1205. doi: 10.3390/biom10091205. Biomolecules. 2020. PMID: 32825095 Free PMC article.
-
A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface.Proc Natl Acad Sci U S A. 2014 Aug 26;111(34):E3506-13. doi: 10.1073/pnas.1412088111. Epub 2014 Aug 13. Proc Natl Acad Sci U S A. 2014. PMID: 25122681 Free PMC article.
-
An Unexpected Encounter: Respiratory Syncytial Virus Nonstructural Protein 1 Interacts with Mediator Subunit MED25.J Virol. 2022 Oct 12;96(19):e0129722. doi: 10.1128/jvi.01297-22. Epub 2022 Sep 14. J Virol. 2022. PMID: 36102648 Free PMC article. Review.
-
The mediator coactivator complex: functional and physical roles in transcriptional regulation.J Cell Sci. 2003 Sep 15;116(Pt 18):3667-75. doi: 10.1242/jcs.00734. J Cell Sci. 2003. PMID: 12917354 Review.
Cited by
-
The Mediator complex as a master regulator of transcription by RNA polymerase II.Nat Rev Mol Cell Biol. 2022 Nov;23(11):732-749. doi: 10.1038/s41580-022-00498-3. Epub 2022 Jun 20. Nat Rev Mol Cell Biol. 2022. PMID: 35725906 Free PMC article. Review.
-
A Lipopeptidomimetic of Transcriptional Activation Domains Selectively Disrupts the Coactivator Med25 Protein-Protein Interactions.Angew Chem Int Ed Engl. 2024 May 21;63(21):e202400781. doi: 10.1002/anie.202400781. Epub 2024 Apr 17. Angew Chem Int Ed Engl. 2024. PMID: 38527936 Free PMC article.
-
αα-Hub domains and intrinsically disordered proteins: A decisive combo.J Biol Chem. 2021 Jan-Jun;296:100226. doi: 10.1074/jbc.REV120.012928. Epub 2020 Dec 29. J Biol Chem. 2021. PMID: 33361159 Free PMC article. Review.
-
Drugging Fuzzy Complexes in Transcription.Front Mol Biosci. 2021 Dec 21;8:795743. doi: 10.3389/fmolb.2021.795743. eCollection 2021. Front Mol Biosci. 2021. PMID: 34993233 Free PMC article.
-
Understanding p300-transcription factor interactions using sequence variation and hybridization.RSC Chem Biol. 2022 Apr 11;3(5):592-603. doi: 10.1039/d2cb00026a. eCollection 2022 May 11. RSC Chem Biol. 2022. PMID: 35656479 Free PMC article.
References
-
- Ma J., Ptashne M., A new class of yeast transcriptional activators. Cell 51, 113–119 (1987). - PubMed
-
- Hope I. A., Mahadevan S., Struhl K., Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature 333, 635–640 (1988). - PubMed
-
- Sigler P. B., Transcriptional activation. Acid blobs and negative noodles. Nature 333, 210–212 (1988). - PubMed
-
- Mapp A. K., Ansari A. Z., A TAD further: Exogenous control of gene activation. ACS Chem. Biol. 2, 62–75 (2007). - PubMed
-
- Piskacek S., et al. ., Nine-amino-acid transactivation domain: Establishment and prediction utilities. Genomics 89, 756–768 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

