cGAS suppresses genomic instability as a decelerator of replication forks
- PMID: 33055160
- PMCID: PMC7556829
- DOI: 10.1126/sciadv.abb8941
cGAS suppresses genomic instability as a decelerator of replication forks
Abstract
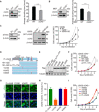
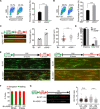
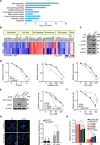
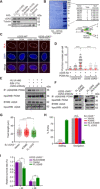
The cyclic GMP-AMP synthase (cGAS), a sensor of cytosolic DNA, is critical for the innate immune response. Here, we show that loss of cGAS in untransformed and cancer cells results in uncontrolled DNA replication, hyperproliferation, and genomic instability. While the majority of cGAS is cytoplasmic, a fraction of cGAS associates with chromatin. cGAS interacts with replication fork proteins in a DNA binding-dependent manner, suggesting that cGAS encounters replication forks in DNA. Independent of cGAMP and STING, cGAS slows replication forks by binding to DNA in the nucleus. In the absence of cGAS, replication forks are accelerated, but fork stability is compromised. Consequently, cGAS-deficient cells are exposed to replication stress and become increasingly sensitive to radiation and chemotherapy. Thus, by acting as a decelerator of DNA replication forks, cGAS controls replication dynamics and suppresses replication-associated DNA damage, suggesting that cGAS is an attractive target for exploiting the genomic instability of cancer cells.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures
Similar articles
-
Cytosolic DNA sensing by cGAS/STING promotes TRPV2-mediated Ca2+ release to protect stressed replication forks.Mol Cell. 2023 Feb 16;83(4):556-573.e7. doi: 10.1016/j.molcel.2022.12.034. Epub 2023 Jan 24. Mol Cell. 2023. PMID: 36696898 Free PMC article.
-
Nuclear cGAS: sequestration and beyond.Protein Cell. 2022 Feb;13(2):90-101. doi: 10.1007/s13238-021-00869-0. Epub 2021 Aug 9. Protein Cell. 2022. PMID: 34374004 Free PMC article. Review.
-
Stalled replication fork protection limits cGAS-STING and P-body-dependent innate immune signalling.Nat Cell Biol. 2022 Jul;24(7):1154-1164. doi: 10.1038/s41556-022-00950-8. Epub 2022 Jul 11. Nat Cell Biol. 2022. PMID: 35817959 Free PMC article.
-
MRE11-dependent instability in mitochondrial DNA fork protection activates a cGAS immune signaling pathway.Sci Adv. 2021 Dec 17;7(51):eabf9441. doi: 10.1126/sciadv.abf9441. Epub 2021 Dec 15. Sci Adv. 2021. PMID: 34910513 Free PMC article.
-
The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer.J Exp Med. 2018 May 7;215(5):1287-1299. doi: 10.1084/jem.20180139. Epub 2018 Apr 5. J Exp Med. 2018. PMID: 29622565 Free PMC article. Review.
Cited by
-
Cyclic GMP-AMP synthase recognizes the physical features of DNA.Acta Pharmacol Sin. 2025 Feb;46(2):264-270. doi: 10.1038/s41401-024-01369-7. Epub 2024 Aug 7. Acta Pharmacol Sin. 2025. PMID: 39112770 Review.
-
Cytosolic DNA sensing by cGAS/STING promotes TRPV2-mediated Ca2+ release to protect stressed replication forks.Mol Cell. 2023 Feb 16;83(4):556-573.e7. doi: 10.1016/j.molcel.2022.12.034. Epub 2023 Jan 24. Mol Cell. 2023. PMID: 36696898 Free PMC article.
-
DNA damage and repair in age-related inflammation.Nat Rev Immunol. 2023 Feb;23(2):75-89. doi: 10.1038/s41577-022-00751-y. Epub 2022 Jul 13. Nat Rev Immunol. 2023. PMID: 35831609 Free PMC article. Review.
-
More than Meets the ISG15: Emerging Roles in the DNA Damage Response and Beyond.Biomolecules. 2020 Nov 15;10(11):1557. doi: 10.3390/biom10111557. Biomolecules. 2020. PMID: 33203188 Free PMC article. Review.
-
The Trinity of cGAS, TLR9, and ALRs Guardians of the Cellular Galaxy Against Host-Derived Self-DNA.Front Immunol. 2021 Feb 11;11:624597. doi: 10.3389/fimmu.2020.624597. eCollection 2020. Front Immunol. 2021. PMID: 33643304 Free PMC article. Review.
References
-
- Pépin G., Gantier M. P., cGAS-STING activation in the tumor microenvironment and its role in cancer immunity. Adv. Exp. Med. Biol. 1024, 175–194 (2017). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

