Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity
- PMID: 33010815
- PMCID: PMC7494270
- DOI: 10.1016/j.cell.2020.09.038
Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity
Abstract
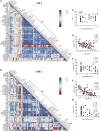
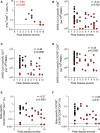
Limited knowledge is available on the relationship between antigen-specific immune responses and COVID-19 disease severity. We completed a combined examination of all three branches of adaptive immunity at the level of SARS-CoV-2-specific CD4+ and CD8+ T cell and neutralizing antibody responses in acute and convalescent subjects. SARS-CoV-2-specific CD4+ and CD8+ T cells were each associated with milder disease. Coordinated SARS-CoV-2-specific adaptive immune responses were associated with milder disease, suggesting roles for both CD4+ and CD8+ T cells in protective immunity in COVID-19. Notably, coordination of SARS-CoV-2 antigen-specific responses was disrupted in individuals ≥ 65 years old. Scarcity of naive T cells was also associated with aging and poor disease outcomes. A parsimonious explanation is that coordinated CD4+ T cell, CD8+ T cell, and antibody responses are protective, but uncoordinated responses frequently fail to control disease, with a connection between aging and impaired adaptive immune responses to SARS-CoV-2.
Keywords: CD4; CD8; CXCL10; IP-10; Spike; T cells; adaptive immunity; antibody; coronavirus; epitopes; neutralizing antibodies.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests A.S. is a consultant for Gritstone, Flow Pharma, and Avalia. S.C. is a consultant for Avalia.
Figures












Similar articles
-
Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19.Nat Immunol. 2020 Nov;21(11):1336-1345. doi: 10.1038/s41590-020-0782-6. Epub 2020 Sep 4. Nat Immunol. 2020. PMID: 32887977 Free PMC article.
-
Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses.Immunity. 2020 Oct 13;53(4):864-877.e5. doi: 10.1016/j.immuni.2020.07.026. Epub 2020 Aug 4. Immunity. 2020. PMID: 32791036 Free PMC article.
-
SARS-CoV-2 T cell immunity: Specificity, function, durability, and role in protection.Sci Immunol. 2020 Jul 17;5(49):eabd6160. doi: 10.1126/sciimmunol.abd6160. Sci Immunol. 2020. PMID: 32680954
-
Remodeling of the Immune Response With Aging: Immunosenescence and Its Potential Impact on COVID-19 Immune Response.Front Immunol. 2020 Aug 7;11:1748. doi: 10.3389/fimmu.2020.01748. eCollection 2020. Front Immunol. 2020. PMID: 32849623 Free PMC article. Review.
-
[Adaptive immunity against SARS-CoV-2].Med Sci (Paris). 2020 Oct;36(10):908-913. doi: 10.1051/medsci/2020168. Epub 2020 Sep 22. Med Sci (Paris). 2020. PMID: 32960167 Review. French.
Cited by
-
Age-dependent immune profile in healthy individuals: an original study, systematic review and meta-analysis.Immun Ageing. 2024 Oct 29;21(1):75. doi: 10.1186/s12979-024-00480-x. Immun Ageing. 2024. PMID: 39472926 Free PMC article.
-
T-Cell Immune Responses to SARS-CoV-2 Infection and Vaccination.Vaccines (Basel). 2024 Sep 30;12(10):1126. doi: 10.3390/vaccines12101126. Vaccines (Basel). 2024. PMID: 39460293 Free PMC article. Review.
-
Activation-Induced Marker Assay to Identify and Isolate HCV-Specific T Cells for Single-Cell RNA-Seq Analysis.Viruses. 2024 Oct 17;16(10):1623. doi: 10.3390/v16101623. Viruses. 2024. PMID: 39459954 Free PMC article.
-
The molecular mechanisms of CD8+ T cell responses to SARS-CoV-2 infection mediated by TCR-pMHC interactions.Front Immunol. 2024 Oct 10;15:1468456. doi: 10.3389/fimmu.2024.1468456. eCollection 2024. Front Immunol. 2024. PMID: 39450171 Free PMC article. Review.
-
SARS-CoV-2-Specific T-Cell as a Potent Therapeutic Strategy against Immune Evasion of Emerging COVID-19 Variants.Int J Mol Sci. 2024 Sep 29;25(19):10512. doi: 10.3390/ijms251910512. Int J Mol Sci. 2024. PMID: 39408840 Free PMC article.
References
-
- Amanna I.J., Slifka M.K., Crotty S. Immunity and immunological memory following smallpox vaccination. Immunol. Rev. 2006;211:320–337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

