Analysis of the Codon Usage Pattern of HA and NA Genes of H7N9 Influenza A Virus
- PMID: 32992529
- PMCID: PMC7583936
- DOI: 10.3390/ijms21197129
Analysis of the Codon Usage Pattern of HA and NA Genes of H7N9 Influenza A Virus
Abstract
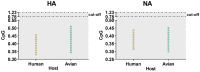
Novel H7N9 influenza virus transmitted from birds to human and, since March 2013, it has caused five epidemic waves in China. Although the evolution of H7N9 viruses has been investigated, the evolutionary changes associated with codon usage are still unclear. Herein, the codon usage pattern of two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), was studied to understand the evolutionary changes in relation to host, epidemic wave, and pathogenicity. Both genes displayed a low codon usage bias, with HA higher than NA. The codon usage was driven by mutation pressure and natural selection, although the main contributing factor was natural selection. Additionally, the codon adaptation index (CAI) and deoptimization (RCDI) illustrated the strong adaptability of H7N9 to Gallus gallus. Similarity index (SiD) analysis showed that Homo sapiens posed a stronger selection pressure than Gallus gallus. Thus, we assume that this may be related to the gradual adaptability of the virus to human. In addition, the host strong selection pressure was validated based on CpG dinucleotide content. In conclusion, this study analyzed the usage of codons of two genes of H7N9 and expanded our understanding of H7N9 host specificity. This aids into the development of control measures against H7N9 influenza virus.
Keywords: H7N9; HA gene; NA gene; codon usage bias; host specificity.
Conflict of interest statement
The author states that the research is in the absence of any competitive interests and conflicts.
Figures


Similar articles
-
The fit of codon usage of human-isolated avian influenza A viruses to human.Infect Genet Evol. 2020 Jul;81:104181. doi: 10.1016/j.meegid.2020.104181. Epub 2020 Jan 7. Infect Genet Evol. 2020. PMID: 31918040
-
Effects of HA and NA glycosylation pattern changes on the transmission of avian influenza A(H7N9) virus in guinea pigs.Biochem Biophys Res Commun. 2016 Oct 14;479(2):192-197. doi: 10.1016/j.bbrc.2016.09.024. Epub 2016 Sep 6. Biochem Biophys Res Commun. 2016. PMID: 27613087
-
[Genetic characteristics of hemagglutinin and neuraminidase of avian influenza A (H7N9) virus in Guizhou province, 2014-2017].Zhonghua Liu Xing Bing Xue Za Zhi. 2018 Nov 10;39(11):1465-1471. doi: 10.3760/cma.j.issn.0254-6450.2018.11.009. Zhonghua Liu Xing Bing Xue Za Zhi. 2018. PMID: 30462955 Chinese.
-
The Effects of Genetic Variation on H7N9 Avian Influenza Virus Pathogenicity.Viruses. 2020 Oct 28;12(11):1220. doi: 10.3390/v12111220. Viruses. 2020. PMID: 33126529 Free PMC article. Review.
-
Enabling the 'host jump': structural determinants of receptor-binding specificity in influenza A viruses.Nat Rev Microbiol. 2014 Dec;12(12):822-31. doi: 10.1038/nrmicro3362. Epub 2014 Nov 10. Nat Rev Microbiol. 2014. PMID: 25383601 Review.
Cited by
-
Codon usage bias analysis of the gene encoding NAD+-dependent DNA ligase protein of Invertebrate iridescent virus 6.Arch Microbiol. 2023 Oct 9;205(11):352. doi: 10.1007/s00203-023-03688-5. Arch Microbiol. 2023. PMID: 37812231
-
Codon Usage for Genetic Diversity, and Evolutionary Dynamics of Novel Porcine Parvoviruses 2 through 7 (PPV2-PPV7).Viruses. 2022 Jan 18;14(2):170. doi: 10.3390/v14020170. Viruses. 2022. PMID: 35215764 Free PMC article.
-
Misclassified: identification of zoonotic transition biomarker candidates for influenza A viruses using deep neural network.Front Genet. 2023 Jul 27;14:1145166. doi: 10.3389/fgene.2023.1145166. eCollection 2023. Front Genet. 2023. PMID: 37576548 Free PMC article.
-
Epidemiology and Evolution of Emerging Porcine Circovirus-like Viruses in Pigs with Hemorrhagic Dysentery and Diarrhea Symptoms in Central China from 2018 to 2021.Viruses. 2021 Nov 15;13(11):2282. doi: 10.3390/v13112282. Viruses. 2021. PMID: 34835090 Free PMC article.
-
Virus Eradication and Synthetic Biology: Changes with SARS-CoV-2?Viruses. 2021 Mar 28;13(4):569. doi: 10.3390/v13040569. Viruses. 2021. PMID: 33800626 Free PMC article.
References
-
- Liu D., Shi W., Shi Y., Wang D., Xiao H., Li W., Bi Y., Wu Y., Li X., Yan J., et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: Phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

