Prospective Longitudinal ctDNA Workflow Reveals Clinically Actionable Alterations in Ovarian Cancer
- PMID: 32914024
- PMCID: PMC7446450
- DOI: 10.1200/PO.18.00343
Prospective Longitudinal ctDNA Workflow Reveals Clinically Actionable Alterations in Ovarian Cancer
Abstract
Purpose: Circulating tumor DNA (ctDNA) detection is a minimally invasive technique that offers dynamic molecular snapshots of genomic alterations in cancer. Although ctDNA markers can be used for early detection of cancers or for monitoring treatment efficacy, the value of ctDNA in guiding treatment decisions in solid cancers is controversial. Here, we monitored ctDNA to detect clinically actionable alterations during treatment of high-grade serous ovarian cancer, the most common and aggressive form of epithelial ovarian cancer with a 5-year survival rate of 43%.
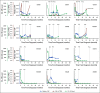
Patients and methods: We implemented a clinical ctDNA workflow to detect clinically actionable alterations in more than 500 cancer-related genes. We applied the workflow to a prospective cohort consisting of 78 ctDNA samples from 12 patients with high-grade serous ovarian cancer before, during, and after treatment. These longitudinal data sets were analyzed using our open-access ctDNA-tailored bioinformatics analysis pipeline and in-house Translational Oncology Knowledgebase to detect clinically actionable genomic alterations. The alterations were ranked according to the European Society for Medical Oncology scale for clinical actionability of molecular targets.
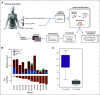
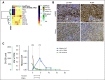
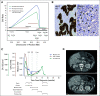
Results: Our results show good concordance of mutations and copy number alterations in ctDNA and tumor samples, and alterations associated with clinically available drugs were detected in seven patients (58%). Treatment of one chemoresistant patient was changed on the basis of detection of ERBB2 amplification, and this ctDNA-guided decision was followed by significant tumor shrinkage and complete normalization of the cancer antigen 125 tumor marker.
Conclusion: Our results demonstrate a proof of concept for using ctDNA to guide clinical decisions. Furthermore, our results show that longitudinal ctDNA samples can be used to identify poor-responding patients after first cycles of chemotherapy. We provide what we believe to be the first comprehensive, open-source ctDNA workflow for detecting clinically actionable alterations in solid cancers.
© 2019 by American Society of Clinical Oncology.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. For a detailed description of the disclosure categories, or for more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifcSakari HietanenConsulting or Advisory Role: Tesaro, AstraZeneca Travel, Accommodations, Expenses: Roche Pharma AGOlli CarpénStock and Other Ownership Interests: Orion Corporation Honoraria: Merck KGaA, Roche Pharma AGSampsa HautaniemiStock and Other Ownership Interests: NanoString Technologies No other potential conflicts of interest were reported.
Figures
Similar articles
-
Extensive mutational ctDNA profiles reflect High-grade serous cancer tumors and reveal emerging mutations at recurrence.Transl Oncol. 2024 Jan;39:101814. doi: 10.1016/j.tranon.2023.101814. Epub 2023 Nov 2. Transl Oncol. 2024. PMID: 37924564 Free PMC article.
-
Correlation of genomic alterations between tumor tissue and circulating tumor DNA by next-generation sequencing.J Cancer Res Clin Oncol. 2018 Nov;144(11):2167-2175. doi: 10.1007/s00432-018-2747-9. Epub 2018 Sep 10. J Cancer Res Clin Oncol. 2018. PMID: 30203147
-
Development and clinical validation of a circulating tumor DNA test for the identification of clinically actionable mutations in nonsmall cell lung cancer.Genes Chromosomes Cancer. 2018 Apr;57(4):211-220. doi: 10.1002/gcc.22522. Epub 2018 Jan 30. Genes Chromosomes Cancer. 2018. PMID: 29277949
-
Dynamic Treatment Stratification Using ctDNA.Recent Results Cancer Res. 2020;215:263-273. doi: 10.1007/978-3-030-26439-0_14. Recent Results Cancer Res. 2020. PMID: 31605234 Review.
-
The Use of Circulating Tumor DNA to Monitor and Predict Response to Treatment in Colorectal Cancer.Front Genet. 2019 Nov 21;10:1118. doi: 10.3389/fgene.2019.01118. eCollection 2019. Front Genet. 2019. PMID: 31824558 Free PMC article. Review.
Cited by
-
Pilot Study of Recurrent Ewing's Sarcoma Management with Vigil/Temozolomide/Irinotecan and Assessment of Circulating Tumor (ct) DNA.Clin Cancer Res. 2023 May 1;29(9):1689-1697. doi: 10.1158/1078-0432.CCR-22-2292. Clin Cancer Res. 2023. PMID: 36780200 Free PMC article.
-
Current Applications and Challenges of Next-Generation Sequencing in Plasma Circulating Tumour DNA of Ovarian Cancer.Biology (Basel). 2024 Jan 31;13(2):88. doi: 10.3390/biology13020088. Biology (Basel). 2024. PMID: 38392306 Free PMC article. Review.
-
Liquid biopsy in ovarian cancer in China and the world: current status and future perspectives.Front Oncol. 2023 Dec 19;13:1276085. doi: 10.3389/fonc.2023.1276085. eCollection 2023. Front Oncol. 2023. PMID: 38169730 Free PMC article. Review.
-
Prognostic Impact of Circulating Methylated Homeobox A9 DNA in Patients Undergoing Treatment for Recurrent Ovarian Cancer.Cancers (Basel). 2022 Mar 30;14(7):1766. doi: 10.3390/cancers14071766. Cancers (Basel). 2022. PMID: 35406538 Free PMC article.
-
Clinical Implication of Liquid Biopsy in Colorectal Cancer Patients Treated with Metastasectomy.Cancers (Basel). 2021 May 6;13(9):2231. doi: 10.3390/cancers13092231. Cancers (Basel). 2021. PMID: 34066481 Free PMC article.
References
-
- Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36:1631–1641. - PubMed
-
- Giannopoulou L, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: Recent advances on circulating tumor cells and circulating tumor DNA. Clin Chem Lab Med. 2018;56:186–197. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

