Deciphering the Molecular Targets and Mechanisms of HGWD in the Treatment of Rheumatoid Arthritis via Network Pharmacology and Molecular Docking
- PMID: 32908565
- PMCID: PMC7471805
- DOI: 10.1155/2020/7151634
Deciphering the Molecular Targets and Mechanisms of HGWD in the Treatment of Rheumatoid Arthritis via Network Pharmacology and Molecular Docking
Abstract
Background: Huangqi Guizhi Wuwu Decoction (HGWD) has been applied in the treatment of joint pain for more than 1000 years in China. Currently, most physicians use HGWD to treat rheumatoid arthritis (RA), and it has proved to have high efficacy. Therefore, it is necessary to explore the potential mechanism of action of HGWD in RA treatment based on network pharmacology and molecular docking methods.
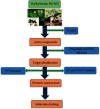
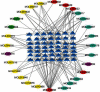
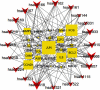
Methods: The active compounds of HGWD were collected, and their targets were identified from the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) and DrugBank database, respectively. The RA-related targets were retrieved by analyzing the differentially expressed genes between RA patients and healthy individuals. Subsequently, the compound-target network of HGWD was constructed and visualized through Cytoscape 3.8.0 software. Protein-protein interaction (PPI) network was constructed to explore the potential mechanisms of HGWD on RA using the plugin BisoGenet of Cytoscape 3.8.0 software. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed in R software (Bioconductor, clusterProfiler). Afterward, molecular docking was used to analyze the binding force of the top 10 active compounds with target proteins of VCAM1, CTNNB1, and JUN.
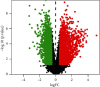
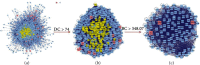
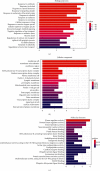
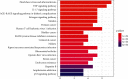
Results: Cumulatively, 790 active compounds and 1006 targets of HGWD were identified. A total of 4570 differentially expressed genes of RA with a p value <0.05 and |log 2(fold change)| > 0.5 were collected. Moreover, 739 GO entries of HGWD on RA were identified, and 79 pathways were screened based on GO and KEGG analysis. The core target gene of HGWD in RA treatment was JUN. Other key target genes included FOS, CCND1, IL6, E2F2, and ICAM1. It was confirmed that the TNF signaling pathway and IL-17 signaling pathway are important pathways of HGWD in the treatment of RA. The molecular docking results revealed that the top 10 active compounds of HGWD had a strong binding to the target proteins of VCAM1, CTNNB1, and JUN.
Conclusion: HGWD has important active compounds such as quercetin, kaempferol, and beta-sitosterol, which exert its therapeutic effect on multiple targets and multiple pathways.
Copyright © 2020 Wei Liu et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

Similar articles
-
Exploring the potential anti-diabetic peripheral neuropathy mechanisms of Huangqi Guizhi Wuwu Decoction by network pharmacology and molecular docking.Metab Brain Dis. 2024 Nov 20;40(1):20. doi: 10.1007/s11011-024-01474-w. Metab Brain Dis. 2024. PMID: 39565454
-
An exploratory study on the mechanism of Huangqi Guizhi Wuwu Decoction in the treatment of neuropathic pain.Ibrain. 2022 May 5;8(2):127-140. doi: 10.1002/ibra.12033. eCollection 2022 Summer. Ibrain. 2022. PMID: 37786887 Free PMC article.
-
The mechanisms of Huangqi Guizhi Wuwu decoction in treating ischaemic stroke based on network pharmacology and experiment verification.Pharm Biol. 2023 Dec;61(1):1014-1029. doi: 10.1080/13880209.2023.2230477. Pharm Biol. 2023. PMID: 37410583 Free PMC article.
-
Exploration of the mechanism of Zisheng Shenqi decoction against gout arthritis using network pharmacology.Comput Biol Chem. 2021 Feb;90:107358. doi: 10.1016/j.compbiolchem.2020.107358. Epub 2020 Aug 8. Comput Biol Chem. 2021. PMID: 33243703 Review.
-
Study on the Efficacy and Safety of the Huangqi Guizhi Wuwu Decoction in the Prevention and Treatment of Chemotherapy-Induced Peripheral Neuropathy: Meta-Analysis of 32 Randomized Controlled Trials.J Pain Res. 2024 Aug 8;17:2605-2628. doi: 10.2147/JPR.S466658. eCollection 2024. J Pain Res. 2024. PMID: 39139997 Free PMC article. Review.
Cited by
-
Integrated Network Pharmacology and Mice Model to Investigate Qing Zao Fang for Treating Sjögren's Syndrome.Evid Based Complement Alternat Med. 2022 Jan 10;2022:3690016. doi: 10.1155/2022/3690016. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 35341135 Free PMC article.
-
The IL-17 family in diseases: from bench to bedside.Signal Transduct Target Ther. 2023 Oct 11;8(1):402. doi: 10.1038/s41392-023-01620-3. Signal Transduct Target Ther. 2023. PMID: 37816755 Free PMC article. Review.
-
Network pharmacology and bioinformatics analysis identified essential genes of Jingulian in the treatment of rheumatoid arthritis and COVID-19.Ann Transl Med. 2022 Jun;10(11):635. doi: 10.21037/atm-22-1665. Ann Transl Med. 2022. PMID: 35813340 Free PMC article.
-
Bioinformatics analysis and experimental validation revealed that Paeoniflorigenone effectively mitigates cerebral ischemic stroke by suppressing oxidative stress and inflammation.Sci Rep. 2024 Mar 7;14(1):5580. doi: 10.1038/s41598-024-55041-5. Sci Rep. 2024. PMID: 38448479 Free PMC article.
-
A study of the therapeutic mechanism of Jakyakgamcho-Tang about functional dyspepsia through network pharmacology research.Int J Med Sci. 2022 Oct 17;19(13):1824-1834. doi: 10.7150/ijms.77451. eCollection 2022. Int J Med Sci. 2022. PMID: 36438925 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

