Cytoplasmic control of intranuclear polarity by human cytomegalovirus
- PMID: 32908309
- PMCID: PMC7644666
- DOI: 10.1038/s41586-020-2714-x
Cytoplasmic control of intranuclear polarity by human cytomegalovirus
Abstract
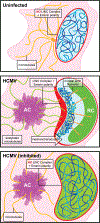
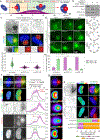
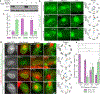
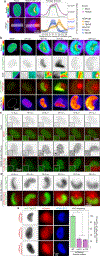
Despite its size and rigidity, the cell nucleus can be moved or reorganized by cytoskeletal filaments under various conditions (for example, during viral infection)1-11. Moreover, whereas chromatin organizes into non-random domains12, extensive heterogeneity at the single-cell level13 means that precisely how and why nuclei reorganize remains an area of intense investigation. Here we describe convolutional neural network-based automated cell classification and analysis pipelines, which revealed the extent to which human cytomegalovirus generates nuclear polarity through a virus-assembled microtubule-organizing centre. Acetylation of tubulin enables microtubules emanating from this centre to rotate the nucleus by engaging cytoplasmically exposed dynein-binding domains in the outer nuclear membrane protein nesprin-2G, which polarizes the inner nuclear membrane protein SUN1. This in turn creates intranuclear polarity in emerin, and thereby controls nuclear actin filaments that spatially segregate viral DNA from inactive histones and host DNA, maximizing virus replication. Our findings demonstrate the extent to which viruses can control the nucleus from the cytoplasm.
Conflict of interest statement
Competing Interest Statement
The authors declare no conflicts of interest associated with this work.
Figures






Similar articles
-
Imbalanced nucleocytoskeletal connections create common polarity defects in progeria and physiological aging.Proc Natl Acad Sci U S A. 2019 Feb 26;116(9):3578-3583. doi: 10.1073/pnas.1809683116. Epub 2019 Feb 11. Proc Natl Acad Sci U S A. 2019. PMID: 30808750 Free PMC article.
-
TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement.J Cell Biol. 2017 Mar 6;216(3):657-674. doi: 10.1083/jcb.201507113. Epub 2017 Feb 27. J Cell Biol. 2017. PMID: 28242745 Free PMC article.
-
The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope.J Cell Sci. 2005 Aug 1;118(Pt 15):3419-30. doi: 10.1242/jcs.02471. J Cell Sci. 2005. PMID: 16079285
-
Nesprin-3: a versatile connector between the nucleus and the cytoskeleton.Biochem Soc Trans. 2011 Dec;39(6):1719-24. doi: 10.1042/BST20110669. Biochem Soc Trans. 2011. PMID: 22103514 Review.
-
Role of cytoskeletal motor proteins in viral infection.Postepy Hig Med Dosw (Online). 2012 Oct 30;66:810-7. doi: 10.5604/17322693.1016360. Postepy Hig Med Dosw (Online). 2012. PMID: 23175336 Review.
Cited by
-
PIP4K2B is mechanoresponsive and controls heterochromatin-driven nuclear softening through UHRF1.Nat Commun. 2023 Mar 14;14(1):1432. doi: 10.1038/s41467-023-37064-0. Nat Commun. 2023. PMID: 36918565 Free PMC article.
-
Negative charge in the RACK1 loop broadens the translational capacity of the human ribosome.Cell Rep. 2021 Sep 7;36(10):109663. doi: 10.1016/j.celrep.2021.109663. Cell Rep. 2021. PMID: 34496247 Free PMC article.
-
mSphere of Influence: Viruses-Pathogens or Expert Cell Biologists?mSphere. 2021 Mar 31;6(2):e00241-21. doi: 10.1128/mSphere.00241-21. mSphere. 2021. PMID: 33789943 Free PMC article.
-
RACK1 Regulates Poxvirus Protein Synthesis Independently of Its Role in Ribosome-Based Stress Signaling.J Virol. 2022 Sep 28;96(18):e0109322. doi: 10.1128/jvi.01093-22. Epub 2022 Sep 13. J Virol. 2022. PMID: 36098514 Free PMC article.
-
Mechanical regulation of chromatin and transcription.Nat Rev Genet. 2022 Oct;23(10):624-643. doi: 10.1038/s41576-022-00493-6. Epub 2022 May 23. Nat Rev Genet. 2022. PMID: 35606569 Review.
References
Methods References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

