LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3-initiated antiviral innate immunity
- PMID: 32907941
- PMCID: PMC7519350
- DOI: 10.1073/pnas.2003932117
LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3-initiated antiviral innate immunity
Abstract
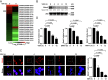
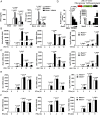
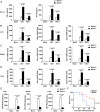
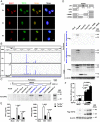
Long noncoding RNAs (lncRNAs) involved in the regulation of antiviral innate immune responses need to be further identified. By functionally screening the lncRNAs in macrophages, here we identified lncRNA Malat1, abundant in the nucleus but significantly down-regulated after viral infection, as a negative regulator of antiviral type I IFN (IFN-I) production. Malat1 directly bound to the transactive response DNA-binding protein (TDP43) in the nucleus and prevented activation of TDP43 by blocking the activated caspase-3-mediated TDP43 cleavage to TDP35. The cleaved TDP35 increased the nuclear IRF3 protein level by binding and degrading Rbck1 pre-mRNA to prevent IRF3 proteasomal degradation upon viral infection, thus selectively promoting antiviral IFN-I production. Deficiency of Malat1 enhanced antiviral innate responses in vivo, accompanying the increased IFN-I production and reduced viral burden. Importantly, the reduced MALAT1, augmented IRF3, and increased IFNA mRNA were found in peripheral blood mononuclear cells (PBMCs) from systemic lupus erythematosus (SLE) patients. Therefore, the down-regulation of MALAT1 in virus-infected cells or in human cells from autoimmune diseases will increase host resistance against viral infection or lead to autoinflammatory interferonopathies via the increased type I IFN production. Our results demonstrate that the nuclear Malat1 suppresses antiviral innate responses by targeting TDP43 activation via RNA-RBP interactive network, adding insight to the molecular regulation of innate responses and autoimmune pathogenesis.
Keywords: Malat1; TDP43; innate immunity; long noncoding RNA; type I interferon.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
MIR155HG Plays a Bivalent Role in Regulating Innate Antiviral Immunity by Encoding Long Noncoding RNA-155 and microRNA-155-5p.mBio. 2022 Dec 20;13(6):e0251022. doi: 10.1128/mbio.02510-22. Epub 2022 Nov 2. mBio. 2022. PMID: 36321836 Free PMC article.
-
The antiviral activity of myricetin against pseudorabies virus through regulation of the type I interferon signaling pathway.J Virol. 2024 Nov 27:e0156724. doi: 10.1128/jvi.01567-24. Online ahead of print. J Virol. 2024. PMID: 39601590
-
SRA Suppresses Antiviral Innate Immune Response in Macrophages by Limiting TBK1 K63 Ubiquitination via Deubiquitinase USP15.Microbiol Spectr. 2022 Dec 21;10(6):e0202822. doi: 10.1128/spectrum.02028-22. Epub 2022 Nov 7. Microbiol Spectr. 2022. PMID: 36342281 Free PMC article.
-
An Emerging Role for Type I Interferons as Critical Regulators of Blood Coagulation.Cells. 2023 Feb 28;12(5):778. doi: 10.3390/cells12050778. Cells. 2023. PMID: 36899914 Free PMC article. Review.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
Cited by
-
LncRNA SNHG3 enhances BMI1 mRNA stability by binding and regulating c-MYC: Implications for the carcinogenic role of SNHG3 in bladder cancer.Cancer Med. 2023 Mar;12(5):5718-5735. doi: 10.1002/cam4.5316. Epub 2022 Oct 8. Cancer Med. 2023. PMID: 36208024 Free PMC article.
-
LncRNA NEAT1 Potentiates SREBP2 Activity to Promote Inflammatory Macrophage Activation and Limit Hantaan Virus Propagation.Front Microbiol. 2022 Apr 13;13:849020. doi: 10.3389/fmicb.2022.849020. eCollection 2022. Front Microbiol. 2022. PMID: 35495674 Free PMC article.
-
LncRNAs, nuclear architecture and the immune response.Nucleus. 2024 Dec;15(1):2350182. doi: 10.1080/19491034.2024.2350182. Epub 2024 May 13. Nucleus. 2024. PMID: 38738760 Free PMC article. Review.
-
Identification and analysis of long non-coding RNAs and mRNAs in chicken macrophages infected with avian infectious bronchitis coronavirus.BMC Genomics. 2021 Jan 20;22(1):67. doi: 10.1186/s12864-020-07359-3. BMC Genomics. 2021. PMID: 33472590 Free PMC article.
-
LNCing RNA to immunity.Trends Immunol. 2022 Jun;43(6):478-495. doi: 10.1016/j.it.2022.04.002. Epub 2022 Apr 29. Trends Immunol. 2022. PMID: 35501219 Free PMC article. Review.
References
-
- Kretschmer S., Lee-Kirsch M. A., Type I interferon-mediated autoinflammation and autoimmunity. Curr. Opin. Immunol. 49, 96–102 (2017). - PubMed
-
- Lee-Kirsch M. A., The type I interferonopathies. Annu. Rev. Med. 68, 297–315 (2017). - PubMed
-
- Liu S. et al. ., Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

