Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19
- PMID: 32887977
- PMCID: PMC7611020
- DOI: 10.1038/s41590-020-0782-6
Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19
Abstract
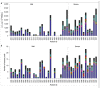
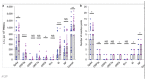
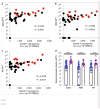
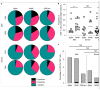
The development of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines and therapeutics will depend on understanding viral immunity. We studied T cell memory in 42 patients following recovery from COVID-19 (28 with mild disease and 14 with severe disease) and 16 unexposed donors, using interferon-γ-based assays with peptides spanning SARS-CoV-2 except ORF1. The breadth and magnitude of T cell responses were significantly higher in severe as compared with mild cases. Total and spike-specific T cell responses correlated with spike-specific antibody responses. We identified 41 peptides containing CD4+ and/or CD8+ epitopes, including six immunodominant regions. Six optimized CD8+ epitopes were defined, with peptide-MHC pentamer-positive cells displaying the central and effector memory phenotype. In mild cases, higher proportions of SARS-CoV-2-specific CD8+ T cells were observed. The identification of T cell responses associated with milder disease will support an understanding of protective immunity and highlights the potential of including non-spike proteins within future COVID-19 vaccine design.
Conflict of interest statement
Figures


Update of
-
Broad and strong memory CD4 + and CD8 + T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients.bioRxiv [Preprint]. 2020 Jun 8:2020.06.05.134551. doi: 10.1101/2020.06.05.134551. bioRxiv. 2020. Update in: Nat Immunol. 2020 Nov;21(11):1336-1345. doi: 10.1038/s41590-020-0782-6. PMID: 32577665 Free PMC article. Updated. Preprint.
Comment in
-
T cells in COVID-19 - united in diversity.Nat Immunol. 2020 Nov;21(11):1307-1308. doi: 10.1038/s41590-020-0798-y. Nat Immunol. 2020. PMID: 32895541 No abstract available.
Similar articles
-
Suboptimal SARS-CoV-2-specific CD8+ T cell response associated with the prominent HLA-A*02:01 phenotype.Proc Natl Acad Sci U S A. 2020 Sep 29;117(39):24384-24391. doi: 10.1073/pnas.2015486117. Epub 2020 Sep 10. Proc Natl Acad Sci U S A. 2020. PMID: 32913053 Free PMC article.
-
Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein.Immunity. 2020 Nov 17;53(5):1095-1107.e3. doi: 10.1016/j.immuni.2020.10.006. Epub 2020 Oct 20. Immunity. 2020. PMID: 33128877 Free PMC article.
-
Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals.Cell. 2020 Jun 25;181(7):1489-1501.e15. doi: 10.1016/j.cell.2020.05.015. Epub 2020 May 20. Cell. 2020. PMID: 32473127 Free PMC article.
-
Amplifying immunogenicity of prospective Covid-19 vaccines by glycoengineering the coronavirus glycan-shield to present α-gal epitopes.Vaccine. 2020 Sep 29;38(42):6487-6499. doi: 10.1016/j.vaccine.2020.08.032. Epub 2020 Aug 19. Vaccine. 2020. PMID: 32907757 Free PMC article. Review.
-
SARS-CoV-2: A New Song Recalls an Old Melody.Cell Host Microbe. 2020 May 13;27(5):692-694. doi: 10.1016/j.chom.2020.04.019. Cell Host Microbe. 2020. PMID: 32407706 Free PMC article. Review.
Cited by
-
High protection and transmission-blocking immunity elicited by single-cycle SARS-CoV-2 vaccine in hamsters.NPJ Vaccines. 2024 Oct 30;9(1):206. doi: 10.1038/s41541-024-00992-z. NPJ Vaccines. 2024. PMID: 39472701 Free PMC article.
-
SARS-CoV-2 variants induce increased inflammatory gene expression but reduced interferon responses and heme synthesis as compared with wild type strains.Sci Rep. 2024 Oct 28;14(1):25734. doi: 10.1038/s41598-024-76401-1. Sci Rep. 2024. PMID: 39468120 Free PMC article.
-
T Cell Peptide Prediction, Immune Response, and Host-Pathogen Relationship in Vaccinated and Recovered from Mild COVID-19 Subjects.Biomolecules. 2024 Sep 26;14(10):1217. doi: 10.3390/biom14101217. Biomolecules. 2024. PMID: 39456150 Free PMC article.
-
The molecular mechanisms of CD8+ T cell responses to SARS-CoV-2 infection mediated by TCR-pMHC interactions.Front Immunol. 2024 Oct 10;15:1468456. doi: 10.3389/fimmu.2024.1468456. eCollection 2024. Front Immunol. 2024. PMID: 39450171 Free PMC article. Review.
-
Allogeneic CD5-specific CAR-T therapy for relapsed/refractory T-ALL: a phase 1 trial.Nat Med. 2024 Oct 1. doi: 10.1038/s41591-024-03282-2. Online ahead of print. Nat Med. 2024. PMID: 39354195
References
Publication types
MeSH terms
Substances
Grants and funding
- 205228/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_19059/MRC_/Medical Research Council/United Kingdom
- G116/150/MRC_/Medical Research Council/United Kingdom
- G1100525/MRC_/Medical Research Council/United Kingdom
- MR/V001329/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_19025/MRC_/Medical Research Council/United Kingdom
- MC_UU_12010/5/MRC_/Medical Research Council/United Kingdom
- MC_UU_00008/6/MRC_/Medical Research Council/United Kingdom
- MC_U137881017/MRC_/Medical Research Council/United Kingdom
- 095541/WT_/Wellcome Trust/United Kingdom
- MC_PC_20002/MRC_/Medical Research Council/United Kingdom
- 215091/WT_/Wellcome Trust/United Kingdom
- MC_UU_00008/11/MRC_/Medical Research Council/United Kingdom
- MC_UU_00008/5/MRC_/Medical Research Council/United Kingdom
- G0501068/MRC_/Medical Research Council/United Kingdom
- MR/N00065X/1/MRC_/Medical Research Council/United Kingdom
- 204969/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 205228/WT_/Wellcome Trust/United Kingdom
- MR/L018942/1/MRC_/Medical Research Council/United Kingdom
- G0600520/MRC_/Medical Research Council/United Kingdom
- G1001046/MRC_/Medical Research Council/United Kingdom
- MR/S032304/1/MRC_/Medical Research Council/United Kingdom
- R01 AI118549/AI/NIAID NIH HHS/United States
- 206377/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 110058/WT_/Wellcome Trust/United Kingdom
- UM1 AI144371/AI/NIAID NIH HHS/United States
- 109965/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 206377/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

