ASTRAL-Pro: Quartet-Based Species-Tree Inference despite Paralogy
- PMID: 32886770
- PMCID: PMC7751180
- DOI: 10.1093/molbev/msaa139
ASTRAL-Pro: Quartet-Based Species-Tree Inference despite Paralogy
Erratum in
-
Corrigendum to: ASTRAL-Pro: Quartet-Based Species-Tree Inference despite Paralogy.Mol Biol Evol. 2021 Sep 27;38(10):4655. doi: 10.1093/molbev/msab232. Mol Biol Evol. 2021. PMID: 34417619 Free PMC article. No abstract available.
Abstract
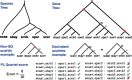
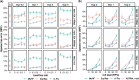
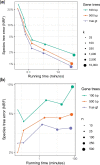
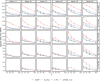
Phylogenetic inference from genome-wide data (phylogenomics) has revolutionized the study of evolution because it enables accounting for discordance among evolutionary histories across the genome. To this end, summary methods have been developed to allow accurate and scalable inference of species trees from gene trees. However, most of these methods, including the widely used ASTRAL, can only handle single-copy gene trees and do not attempt to model gene duplication and gene loss. As a result, most phylogenomic studies have focused on single-copy genes and have discarded large parts of the data. Here, we first propose a measure of quartet similarity between single-copy and multicopy trees that accounts for orthology and paralogy. We then introduce a method called ASTRAL-Pro (ASTRAL for PaRalogs and Orthologs) to find the species tree that optimizes our quartet similarity measure using dynamic programing. By studying its performance on an extensive collection of simulated data sets and on real data sets, we show that ASTRAL-Pro is more accurate than alternative methods.
Keywords: gene duplication and loss; incomplete lineage sorting; species-tree inference.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Similar articles
-
ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees.BMC Bioinformatics. 2018 May 8;19(Suppl 6):153. doi: 10.1186/s12859-018-2129-y. BMC Bioinformatics. 2018. PMID: 29745866 Free PMC article.
-
DISCO: Species Tree Inference using Multicopy Gene Family Tree Decomposition.Syst Biol. 2022 Apr 19;71(3):610-629. doi: 10.1093/sysbio/syab070. Syst Biol. 2022. PMID: 34450658 Free PMC article.
-
Multi-allele species reconstruction using ASTRAL.Mol Phylogenet Evol. 2019 Jan;130:286-296. doi: 10.1016/j.ympev.2018.10.033. Epub 2018 Oct 26. Mol Phylogenet Evol. 2019. PMID: 30393186
-
Inferring orthology and paralogy.Methods Mol Biol. 2012;855:259-79. doi: 10.1007/978-1-61779-582-4_9. Methods Mol Biol. 2012. PMID: 22407712 Review.
-
The inference of gene trees with species trees.Syst Biol. 2015 Jan;64(1):e42-62. doi: 10.1093/sysbio/syu048. Epub 2014 Jul 28. Syst Biol. 2015. PMID: 25070970 Free PMC article. Review.
Cited by
-
Challenges in Assembling the Dated Tree of Life.Genome Biol Evol. 2024 Oct 9;16(10):evae229. doi: 10.1093/gbe/evae229. Genome Biol Evol. 2024. PMID: 39475308 Free PMC article.
-
Dollo-CDP: a polynomial-time algorithm for the clade-constrained large Dollo parsimony problem.Algorithms Mol Biol. 2024 Jan 8;19(1):2. doi: 10.1186/s13015-023-00249-9. Algorithms Mol Biol. 2024. PMID: 38191515 Free PMC article.
-
Analysis of Paralogs in Target Enrichment Data Pinpoints Multiple Ancient Polyploidy Events in Alchemilla s.l. (Rosaceae).Syst Biol. 2021 Dec 16;71(1):190-207. doi: 10.1093/sysbio/syab032. Syst Biol. 2021. PMID: 33978764 Free PMC article.
-
Gapless genome assembly of azalea and multi-omics investigation into divergence between two species with distinct flower color.Hortic Res. 2022 Oct 26;10(1):uhac241. doi: 10.1093/hr/uhac241. eCollection 2023. Hortic Res. 2022. PMID: 36643737 Free PMC article.
-
Phylotranscriptomics Resolves the Phylogeny of Pooideae and Uncovers Factors for Their Adaptive Evolution.Mol Biol Evol. 2022 Feb 3;39(2):msac026. doi: 10.1093/molbev/msac026. Mol Biol Evol. 2022. PMID: 35134207 Free PMC article.
References
-
- An J, Zhu L, Zhang Y, Tang H.. 2013. Efficient visible light photo-fenton-like degradation of organic pollutants using in situ surface-modified BiFeO3 as a catalyst. J Environ Sci (China )25(6):1213–1225. - PubMed
-
- Arvestad L, Berglund A-C, Lagergren J, Sennblad B.. 2004. Gene tree reconstruction and orthology analysis based on an integrated model for duplications and sequence evolution. In: Proceedings of the eighth annual international conference on computational molecular biology—RECOMB ’04; New York: ACM Press. p. 326–335.
-
- Arvestad L, Lagergren J, Sennblad B.. 2009. The gene evolution model and computing its associated probabilities. J ACM 56(2):1–44.
-
- Ballesteros JA, Hormiga G.. 2016. A new orthology assessment method for phylogenomic data: unrooted phylogenetic orthology. Mol Biol Evol. 33(8):2117–2134. - PubMed
-
- Ballesteros JA, Sharma PP.. 2019. A critical appraisal of the placement of Xiphosura (Chelicerata) with account of known sources of phylogenetic error. Syst Biol. 68(6):896–862. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

