Gut dysbiosis contributes to amyloid pathology, associated with C/EBPβ/AEP signaling activation in Alzheimer's disease mouse model
- PMID: 32832679
- PMCID: PMC7439296
- DOI: 10.1126/sciadv.aba0466
Gut dysbiosis contributes to amyloid pathology, associated with C/EBPβ/AEP signaling activation in Alzheimer's disease mouse model
Abstract
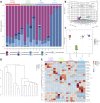
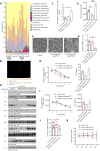
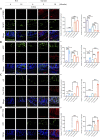
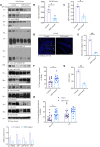
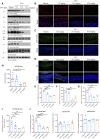
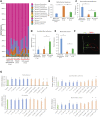
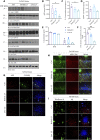
The gut-brain axis is bidirectional, and gut microbiota influence brain disorders including Alzheimer's disease (AD). CCAAT/enhancer binding protein β/asparagine endopeptidase (C/EBPβ/AEP) signaling spatiotemporally mediates AD pathologies in the brain via cleaving both β-amyloid precursor protein and Tau. We show that gut dysbiosis occurs in 5xFAD mice, and is associated with escalation of the C/EBPβ/AEP pathway in the gut with age. Unlike that of aged wild-type mice, the microbiota of aged 3xTg mice accelerate AD pathology in young 3xTg mice, accompanied by active C/EBPβ/AEP signaling in the brain. Antibiotic treatment diminishes this signaling and attenuates amyloidogenic processes in 5xFAD, improving cognitive functions. The prebiotic R13 inhibits this pathway and suppresses amyloid aggregates in the gut. R13-induced Lactobacillus salivarius antagonizes the C/EBPβ/AEP axis, mitigating gut leakage and oxidative stress. Our findings support the hypothesis that C/EBPβ/AEP signaling is activated by gut dysbiosis, implicated in AD pathologies in the gut.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures
Similar articles
-
Patchouli alcohol attenuates the cognitive deficits in a transgenic mouse model of Alzheimer's disease via modulating neuropathology and gut microbiota through suppressing C/EBPβ/AEP pathway.J Neuroinflammation. 2023 Jan 30;20(1):19. doi: 10.1186/s12974-023-02704-1. J Neuroinflammation. 2023. PMID: 36717922 Free PMC article.
-
Gut inflammation triggers C/EBPβ/δ-secretase-dependent gut-to-brain propagation of Aβ and Tau fibrils in Alzheimer's disease.EMBO J. 2021 Sep 1;40(17):e106320. doi: 10.15252/embj.2020106320. Epub 2021 Jul 14. EMBO J. 2021. PMID: 34260075 Free PMC article.
-
Spatiotemporal activation of the C/EBPβ/δ-secretase axis regulates the pathogenesis of Alzheimer's disease.Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):E12427-E12434. doi: 10.1073/pnas.1815915115. Epub 2018 Dec 10. Proc Natl Acad Sci U S A. 2018. PMID: 30530690 Free PMC article.
-
C/EBPβ/AEP Signaling Drives Alzheimer's Disease Pathogenesis.Neurosci Bull. 2023 Jul;39(7):1173-1185. doi: 10.1007/s12264-023-01025-w. Epub 2023 Feb 3. Neurosci Bull. 2023. PMID: 36735152 Free PMC article. Review.
-
Role of gut-brain axis, gut microbial composition, and probiotic intervention in Alzheimer's disease.Life Sci. 2021 Jan 1;264:118627. doi: 10.1016/j.lfs.2020.118627. Epub 2020 Oct 22. Life Sci. 2021. PMID: 33169684 Review.
Cited by
-
GLP-1 Analogs, SGLT-2, and DPP-4 Inhibitors: A Triad of Hope for Alzheimer's Disease Therapy.Biomedicines. 2023 Nov 12;11(11):3035. doi: 10.3390/biomedicines11113035. Biomedicines. 2023. PMID: 38002034 Free PMC article. Review.
-
Sodium oligomannate disrupts the adherence of Ribhigh bacteria to gut epithelia to block SAA-triggered Th1 inflammation in 5XFAD transgenic mice.Cell Discov. 2024 Nov 19;10(1):115. doi: 10.1038/s41421-024-00725-5. Cell Discov. 2024. PMID: 39557828 Free PMC article.
-
Inhibition of microfold cells ameliorates early pathological phenotypes by modulating microglial functions in Alzheimer's disease mouse model.J Neuroinflammation. 2023 Nov 27;20(1):282. doi: 10.1186/s12974-023-02966-9. J Neuroinflammation. 2023. PMID: 38012646 Free PMC article.
-
The link between gut microbiome and Alzheimer's disease: From the perspective of new revised criteria for diagnosis and staging of Alzheimer's disease.Alzheimers Dement. 2024 Aug;20(8):5771-5788. doi: 10.1002/alz.14057. Epub 2024 Jun 28. Alzheimers Dement. 2024. PMID: 38940631 Free PMC article. Review.
-
The role of the gut microbiota in neurodegenerative diseases targeting metabolism.Front Neurosci. 2024 Sep 26;18:1432659. doi: 10.3389/fnins.2024.1432659. eCollection 2024. Front Neurosci. 2024. PMID: 39391755 Free PMC article. Review.
References
-
- Heneka M. T., Carson M. J., Khoury J. E., Landreth G. E., Brosseron F., Feinstein D. L., Jacobs A. H., Wyss-Coray T., Vitorica J., Ransohoff R. M., Herrup K., Frautschy S. A., Finsen B., Brown G. C., Verkhratsky A., Yamanaka K., Koistinaho J., Latz E., Halle A., Petzold G. C., Town T., Morgan D., Shinohara M. L., Perry V. H., Holmes C., Bazan N. G., Brooks D. J., Hunot S., Joseph B., Deigendesch N., Garaschuk O., Boddeke E., Dinarello C. A., Breitner J. C., Cole G. M., Golenbock D. T., Kummer M. P., Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14, 388–405 (2015). - PMC - PubMed
-
- Hu X., Wang T., Jin F., Alzheimer's disease and gut microbiota. Sci. China Life Sci. 59, 1006–1023 (2016). - PubMed
-
- Zhuang Z. Q., Shen L.-L., Li W.-W., Fu X., Zeng F., Gui L., Lü Y., Cai M., Zhu C., Tan Y.-L., Zheng P., Li H.-Y., Zhu J., Zhou H.-D., Bu X.-L., Wang Y.-J., Gut microbiota is altered in patients with Alzheimer’s disease. J. Alzheimers Dis. 63, 1337–1346 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

