The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity
- PMID: 32730807
- PMCID: PMC7366990
- DOI: 10.1016/j.cell.2020.07.012
The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity
Abstract
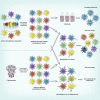
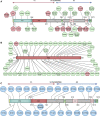
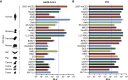
The spike protein of SARS-CoV-2 has been undergoing mutations and is highly glycosylated. It is critically important to investigate the biological significance of these mutations. Here, we investigated 80 variants and 26 glycosylation site modifications for the infectivity and reactivity to a panel of neutralizing antibodies and sera from convalescent patients. D614G, along with several variants containing both D614G and another amino acid change, were significantly more infectious. Most variants with amino acid change at receptor binding domain were less infectious, but variants including A475V, L452R, V483A, and F490L became resistant to some neutralizing antibodies. Moreover, the majority of glycosylation deletions were less infectious, whereas deletion of both N331 and N343 glycosylation drastically reduced infectivity, revealing the importance of glycosylation for viral infectivity. Interestingly, N234Q was markedly resistant to neutralizing antibodies, whereas N165Q became more sensitive. These findings could be of value in the development of vaccine and therapeutic antibodies.
Keywords: COVID-19; SARS-CoV-2; glycosylation; mutants; pseudotyped virus; spike; variants.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



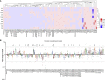

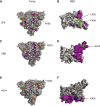
Comment in
-
Profiling and characterization of SARS-CoV-2 mutants' infectivity and antigenicity.Signal Transduct Target Ther. 2020 Sep 3;5(1):185. doi: 10.1038/s41392-020-00302-8. Signal Transduct Target Ther. 2020. PMID: 32883949 Free PMC article. No abstract available.
-
Cellular tropism and antigenicity of mink-derived SARS-CoV-2 variants.Signal Transduct Target Ther. 2021 May 17;6(1):196. doi: 10.1038/s41392-021-00617-0. Signal Transduct Target Ther. 2021. PMID: 34001855 Free PMC article. No abstract available.
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Safety and immunogenicity of an egg-based inactivated Newcastle disease virus vaccine expressing SARS-CoV-2 spike: Interim results of a randomized, placebo-controlled, phase 1/2 trial in Vietnam.Vaccine. 2022 Jun 9;40(26):3621-3632. doi: 10.1016/j.vaccine.2022.04.078. Epub 2022 May 14. Vaccine. 2022. PMID: 35577631 Free PMC article. Clinical Trial.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity.Nature. 2022 Mar;603(7902):706-714. doi: 10.1038/s41586-022-04474-x. Epub 2022 Feb 1. Nature. 2022. PMID: 35104837 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
Neutralizing antibodies against SARS-CoV-2: current understanding, challenge and perspective.Antib Ther. 2020 Dec 28;3(4):285-299. doi: 10.1093/abt/tbaa028. eCollection 2020 Dec. Antib Ther. 2020. PMID: 33912797 Free PMC article. Review.
-
Structural impact on SARS-CoV-2 spike protein by D614G substitution.Science. 2021 Apr 30;372(6541):525-530. doi: 10.1126/science.abf2303. Epub 2021 Mar 16. Science. 2021. PMID: 33727252 Free PMC article.
-
Deletion of the SARS-CoV-2 Spike Cytoplasmic Tail Increases Infectivity in Pseudovirus Neutralization Assays.J Virol. 2021 May 10;95(11):e00044-21. doi: 10.1128/JVI.00044-21. Epub 2021 Mar 16. J Virol. 2021. PMID: 33727331 Free PMC article.
-
A human antibody of potent efficacy against SARS-CoV-2 in rhesus macaques showed strong blocking activity to B.1.351.MAbs. 2021 Jan-Dec;13(1):1930636. doi: 10.1080/19420862.2021.1930636. MAbs. 2021. PMID: 34097570 Free PMC article.
-
Mechanosensing view of SARS-CoV-2 infection by a DNA nano-assembly.Cell Rep Phys Sci. 2022 Sep 21;3(9):101048. doi: 10.1016/j.xcrp.2022.101048. Cell Rep Phys Sci. 2022. PMID: 36157982 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

