Molecular mechanisms underlying menthol binding and activation of TRPM8 ion channel
- PMID: 32728032
- PMCID: PMC7391767
- DOI: 10.1038/s41467-020-17582-x
Molecular mechanisms underlying menthol binding and activation of TRPM8 ion channel
Abstract
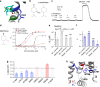
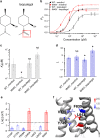
Menthol in mints elicits coolness sensation by selectively activating TRPM8 channel. Although structures of TRPM8 were determined in the apo and liganded states, the menthol-bounded state is unresolved. To understand how menthol activates the channel, we docked menthol to the channel and systematically validated our menthol binding models with thermodynamic mutant cycle analysis. We observed that menthol uses its hydroxyl group as a hand to specifically grab with R842, and its isopropyl group as legs to stand on I846 and L843. By imaging with fluorescent unnatural amino acid, we found that menthol binding induces wide-spread conformational rearrangements within the transmembrane domains. By Φ analysis based on single-channel recordings, we observed a temporal sequence of conformational changes in the S6 bundle crossing and the selectivity filter leading to channel activation. Therefore, our study suggested a 'grab and stand' mechanism of menthol binding and how menthol activates TRPM8 at the atomic level.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Implications of Human Transient Receptor Potential Melastatin 8 (TRPM8) Channel Gating from Menthol Binding Studies of the Sensing Domain.Biochemistry. 2016 Jan 12;55(1):114-24. doi: 10.1021/acs.biochem.5b00931. Epub 2015 Dec 23. Biochemistry. 2016. PMID: 26653082 Free PMC article.
-
Bidirectional modulation of thermal and chemical sensitivity of TRPM8 channels by the initial region of the N-terminal domain.J Biol Chem. 2014 Aug 8;289(32):21828-43. doi: 10.1074/jbc.M114.565994. Epub 2014 Jun 10. J Biol Chem. 2014. PMID: 24917670 Free PMC article.
-
Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP2.Science. 2022 Oct 14;378(6616):eadd1268. doi: 10.1126/science.add1268. Epub 2022 Oct 14. Science. 2022. PMID: 36227998 Free PMC article.
-
Mutations of TRPM8 channels: Unraveling the molecular basis of activation by cold and ligands.Med Res Rev. 2022 Nov;42(6):2168-2203. doi: 10.1002/med.21920. Epub 2022 Aug 17. Med Res Rev. 2022. PMID: 35976012 Free PMC article. Review.
-
TRPM8 ion channel ligands for new therapeutic applications and as probes to study menthol pharmacology.Life Sci. 2013 Mar 19;92(8-9):425-37. doi: 10.1016/j.lfs.2012.10.032. Epub 2012 Nov 16. Life Sci. 2013. PMID: 23159643 Review.
Cited by
-
The distinctive role of menthol in pain and analgesia: Mechanisms, practices, and advances.Front Mol Neurosci. 2022 Oct 5;15:1006908. doi: 10.3389/fnmol.2022.1006908. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36277488 Free PMC article. Review.
-
Investigating the role of nicotinic acetylcholine receptors in menthol's effects in mice.Drug Alcohol Depend. 2024 Apr 1;257:111262. doi: 10.1016/j.drugalcdep.2024.111262. Epub 2024 Mar 8. Drug Alcohol Depend. 2024. PMID: 38492255
-
Cold stimuli, hot topic: An updated review on the biological activity of menthol in relation to inflammation.Front Immunol. 2022 Nov 9;13:1023746. doi: 10.3389/fimmu.2022.1023746. eCollection 2022. Front Immunol. 2022. PMID: 36439160 Free PMC article. Review.
-
Rational Design of a Modality-Specific Inhibitor of TRPM8 Channel against Oxaliplatin-Induced Cold Allodynia.Adv Sci (Weinh). 2021 Nov;8(22):e2101717. doi: 10.1002/advs.202101717. Epub 2021 Oct 17. Adv Sci (Weinh). 2021. PMID: 34658162 Free PMC article.
-
Nebivolol as a Potent TRPM8 Channel Blocker: A Drug-Screening Approach through Automated Patch Clamping and Ligand-Based Virtual Screening.Membranes (Basel). 2022 Sep 28;12(10):954. doi: 10.3390/membranes12100954. Membranes (Basel). 2022. PMID: 36295712 Free PMC article.
References
-
- Galeotti N, Di Cesare Mannelli L, Mazzanti G, Bartolini A, Ghelardini C. Menthol: a natural analgesic compound. Neurosci. Lett. 2002;322:145–148. - PubMed
-
- Stander S, et al. Novel TRPM8 agonist cooling compound against chronic itch: results from a randomized, double-blind, controlled, pilot study in dry skin. J. Eur. Acad. Dermatol. Venereol. 2017;31:1064–1068. - PubMed
-
- Peier AM, et al. A TRP channel that senses cold stimuli and menthol. Cell. 2002;108:705–715. - PubMed
-
- McKemy DD, Neuhausser WM, Julius D. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature. 2002;416:52–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

