Mitochondria and Peroxisome Remodeling across Cytomegalovirus Infection Time Viewed through the Lens of Inter-ViSTA
- PMID: 32726614
- PMCID: PMC8489195
- DOI: 10.1016/j.celrep.2020.107943
Mitochondria and Peroxisome Remodeling across Cytomegalovirus Infection Time Viewed through the Lens of Inter-ViSTA
Abstract
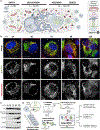
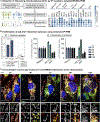
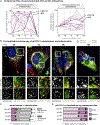
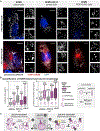
Nearly all biological processes rely on the finely tuned coordination of protein interactions across cellular space and time. Accordingly, generating protein interactomes has become routine in biological studies, yet interpreting these datasets remains computationally challenging. Here, we introduce Inter-ViSTA (Interaction Visualization in Space and Time Analysis), a web-based platform that quickly builds animated protein interaction networks and automatically synthesizes information on protein abundances, functions, complexes, and subcellular localizations. Using Inter-ViSTA with proteomics and molecular virology, we define virus-host interactions for the human cytomegalovirus (HCMV) anti-apoptotic protein, pUL37x1. We find that spatiotemporal controlled interactions underlie pUL37x1 functions, facilitating the pro-viral remodeling of mitochondria and peroxisomes during infection. Reciprocal isolations, microscopy, and genetic manipulations further characterize these associations, revealing the interplay between pUL37x1 and the MIB complex, which is critical for mitochondrial integrity. At the peroxisome, we show that pUL37x1 activates PEX11β to regulate fission, a key aspect of virus assembly and spread.
Keywords: HCMV; IP-MS; Inter-ViSTA; MICOS; mitochondria; pUL37; peroxisome; protein interactions; vMIA; virus-host interactions.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



Similar articles
-
The human cytomegalovirus protein UL37 exon 1 associates with internal lipid rafts.J Virol. 2011 Mar;85(5):2100-11. doi: 10.1128/JVI.01830-10. Epub 2010 Dec 22. J Virol. 2011. PMID: 21177823 Free PMC article.
-
HCMV strain- and cell type-specific alterations in membrane contact sites point to the convergent regulation of organelle remodeling.J Virol. 2024 Nov 19;98(11):e0109924. doi: 10.1128/jvi.01099-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480111
-
Mitochondrial and secretory human cytomegalovirus UL37 proteins traffic into mitochondrion-associated membranes of human cells.J Virol. 2008 Mar;82(6):2715-26. doi: 10.1128/JVI.02456-07. Epub 2008 Jan 16. J Virol. 2008. PMID: 18199645 Free PMC article.
-
vMIA, a viral inhibitor of apoptosis targeting mitochondria.Biochimie. 2002 Feb-Mar;84(2-3):177-85. doi: 10.1016/s0300-9084(02)01367-6. Biochimie. 2002. PMID: 12022948 Review.
-
Superresolution imaging of viral protein trafficking.Med Microbiol Immunol. 2015 Jun;204(3):449-60. doi: 10.1007/s00430-015-0395-0. Epub 2015 Feb 28. Med Microbiol Immunol. 2015. PMID: 25724304 Free PMC article. Review.
Cited by
-
Restructured membrane contacts rewire organelles for human cytomegalovirus infection.Nat Commun. 2022 Aug 11;13(1):4720. doi: 10.1038/s41467-022-32488-6. Nat Commun. 2022. PMID: 35953480 Free PMC article.
-
The roles of autophagy and mitophagy in corneal pathology: current knowledge and future perspectives.Front Med (Lausanne). 2023 Apr 21;10:1064938. doi: 10.3389/fmed.2023.1064938. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37153108 Free PMC article. Review.
-
Targeting the Host Mitochondria as a Novel Human Cytomegalovirus Antiviral Strategy.Viruses. 2023 Apr 28;15(5):1083. doi: 10.3390/v15051083. Viruses. 2023. PMID: 37243170 Free PMC article. Review.
-
The human cytomegalovirus protein pUL13 targets mitochondrial cristae architecture to increase cellular respiration during infection.Proc Natl Acad Sci U S A. 2021 Aug 10;118(32):e2101675118. doi: 10.1073/pnas.2101675118. Proc Natl Acad Sci U S A. 2021. PMID: 34344827 Free PMC article.
-
Human Cytomegalovirus Modifies Placental Small Extracellular Vesicle Composition to Enhance Infection of Fetal Neural Cells In Vitro.Viruses. 2022 Sep 13;14(9):2030. doi: 10.3390/v14092030. Viruses. 2022. PMID: 36146834 Free PMC article.
References
-
- Alexa A, and Rahnenführer J. (2018). Gene set enrichment analysis with topGO.
-
- Bender-deMoll S. (2016). Temporal network tools in statnet: networkDynamic, ndtv and tsna. https://statnet.org/Workshops/ndtv_workshop.html.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

