Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein
- PMID: 32723359
- PMCID: PMC7386171
- DOI: 10.1186/s13073-020-00763-0
Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein
Abstract
Background: SARS-CoV-2 is a recently emerged respiratory pathogen that has significantly impacted global human health. We wanted to rapidly characterise the transcriptomic, proteomic and phosphoproteomic landscape of this novel coronavirus to provide a fundamental description of the virus's genomic and proteomic potential.
Methods: We used direct RNA sequencing to determine the transcriptome of SARS-CoV-2 grown in Vero E6 cells which is widely used to propagate the novel coronavirus. The viral transcriptome was analysed using a recently developed ORF-centric pipeline. Allied to this, we used tandem mass spectrometry to investigate the proteome and phosphoproteome of the same virally infected cells.
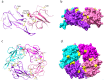
Results: Our integrated analysis revealed that the viral transcripts (i.e. subgenomic mRNAs) generally fitted the expected transcription model for coronaviruses. Importantly, a 24 nt in-frame deletion was detected in over half of the subgenomic mRNAs encoding the spike (S) glycoprotein and was predicted to remove a proposed furin cleavage site from the S glycoprotein. Tandem mass spectrometry identified over 500 viral peptides and 44 phosphopeptides in virus-infected cells, covering almost all proteins predicted to be encoded by the SARS-CoV-2 genome, including peptides unique to the deleted variant of the S glycoprotein.
Conclusions: Detection of an apparently viable deletion in the furin cleavage site of the S glycoprotein, a leading vaccine target, shows that this and other regions of SARS-CoV-2 proteins may readily mutate. The furin site directs cleavage of the S glycoprotein into functional subunits during virus entry or exit and likely contributes strongly to the pathogenesis and zoonosis of this virus. Our data emphasises that the viral genome sequence should be carefully monitored during the growth of viral stocks for research, animal challenge models and, potentially, in clinical samples. Such variations may result in different levels of virulence, morbidity and mortality.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


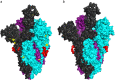

Similar articles
-
Identification of Common Deletions in the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2.J Virol. 2020 Aug 17;94(17):e00790-20. doi: 10.1128/JVI.00790-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32571797 Free PMC article.
-
The SARS-CoV-2 Transcriptome and the Dynamics of the S Gene Furin Cleavage Site in Primary Human Airway Epithelia.mBio. 2021 May 11;12(3):e01006-21. doi: 10.1128/mBio.01006-21. mBio. 2021. PMID: 33975939 Free PMC article.
-
SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients.J Gen Virol. 2020 Nov;101(11):1156-1169. doi: 10.1099/jgv.0.001481. Epub 2020 Aug 21. J Gen Virol. 2020. PMID: 32821033 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Unraveling the Epidemiology, Geographical Distribution, and Genomic Evolution of Potentially Lethal Coronaviruses (SARS, MERS, and SARS CoV-2).Front Cell Infect Microbiol. 2020 Aug 27;10:499. doi: 10.3389/fcimb.2020.00499. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32974224 Free PMC article. Review.
Cited by
-
Interaction between Sars-CoV-2 structural proteins and host cellular receptors: From basic mechanisms to clinical perspectives.Adv Protein Chem Struct Biol. 2022;132:243-277. doi: 10.1016/bs.apcsb.2022.05.010. Epub 2022 Jun 9. Adv Protein Chem Struct Biol. 2022. PMID: 36088078 Free PMC article. Review.
-
Sequential Infection with Influenza A Virus Followed by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Leads to More Severe Disease and Encephalitis in a Mouse Model of COVID-19.Viruses. 2024 May 28;16(6):863. doi: 10.3390/v16060863. Viruses. 2024. PMID: 38932155 Free PMC article.
-
SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes.bioRxiv [Preprint]. 2020 Sep 2:2020.06.02.130955. doi: 10.1101/2020.06.02.130955. bioRxiv. 2020. Update in: Nat Commun. 2021 May 11;12(1):2642. doi: 10.1038/s41467-021-22905-7. PMID: 32577641 Free PMC article. Updated. Preprint.
-
Identification of Hammerhead-variant ribozyme sequences in SARS-CoV-2.Nucleic Acids Res. 2024 Apr 12;52(6):3262-3277. doi: 10.1093/nar/gkae037. Nucleic Acids Res. 2024. PMID: 38296822 Free PMC article.
-
COVID-19: from rapid genome sequencing to fast decisions.Lancet Infect Dis. 2020 Nov;20(11):1218. doi: 10.1016/S1473-3099(20)30580-6. Epub 2020 Jul 14. Lancet Infect Dis. 2020. PMID: 32679087 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

