FOXO transcription factors activate alternative major immediate early promoters to induce human cytomegalovirus reactivation
- PMID: 32694203
- PMCID: PMC7414233
- DOI: 10.1073/pnas.2002651117
FOXO transcription factors activate alternative major immediate early promoters to induce human cytomegalovirus reactivation
Abstract
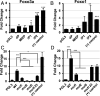
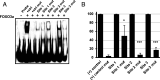
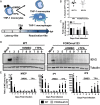
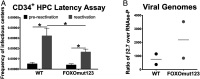
Human progenitor cells (HPCs) support human cytomegalovirus (HCMV) latency, and their differentiation along the myeloid lineage triggers cellular cues that drive reactivation. A key step during HCMV reactivation in latently infected HPCs is reexpression of viral major immediate early (MIE) genes. We recently determined that the major immediate early promoter (MIEP), which is primarily responsible for MIE gene expression during lytic replication, remains silent during reactivation. Instead, alternative promoters in the MIE locus are induced by reactivation stimuli. Here, we find that forkhead family (FOXO) transcription factors are critical for activation of alternative MIE promoters during HCMV reactivation, as mutating FOXO binding sites in alternative MIE promoters decreased HCMV IE gene expression upon reactivation and significantly decreased the production of infectious virus from latently infected primary CD34+ HPCs. These findings establish a mechanistic link by which infected cells sense environmental cues to regulate latency and reactivation, and emphasize the role of contextual activation of alternative MIE promoters as the primary drivers of reactivation.
Keywords: FOXO; HCMV; herpesvirus; latency; reactivation.
Conflict of interest statement
Competing interest statement: J.P.K., F.G., and N.J.M. have filed a patent for manipulation of intronic promoters to reduce reactivation of viral vaccines.
Figures
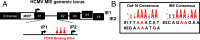

Similar articles
-
Control of Immediate Early Gene Expression for Human Cytomegalovirus Reactivation.Front Cell Infect Microbiol. 2020 Sep 17;10:476. doi: 10.3389/fcimb.2020.00476. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33072616 Free PMC article. Review.
-
Activator protein-1 transactivation of the major immediate early locus is a determinant of cytomegalovirus reactivation from latency.Proc Natl Acad Sci U S A. 2020 Aug 25;117(34):20860-20867. doi: 10.1073/pnas.2009420117. Epub 2020 Aug 11. Proc Natl Acad Sci U S A. 2020. PMID: 32788362 Free PMC article.
-
Human cytomegalovirus major immediate early transcripts arise predominantly from the canonical major immediate early promoter in reactivating progenitor-derived dendritic cells.J Gen Virol. 2020 Jun;101(6):635-644. doi: 10.1099/jgv.0.001419. J Gen Virol. 2020. PMID: 32375946 Free PMC article.
-
Alternative promoters drive human cytomegalovirus reactivation from latency.Proc Natl Acad Sci U S A. 2019 Aug 27;116(35):17492-17497. doi: 10.1073/pnas.1900783116. Epub 2019 Aug 13. Proc Natl Acad Sci U S A. 2019. PMID: 31409717 Free PMC article.
-
Human cytomegalovirus: Latency and reactivation in the myeloid lineage.J Clin Virol. 2008 Mar;41(3):180-5. doi: 10.1016/j.jcv.2007.11.014. J Clin Virol. 2008. PMID: 18164651 Review.
Cited by
-
Virus-induced FoxO factor facilitates replication of human cytomegalovirus.Arch Virol. 2022 Jan;167(1):109-121. doi: 10.1007/s00705-021-05279-5. Epub 2021 Nov 9. Arch Virol. 2022. PMID: 34751815
-
The Virus-Induced Upregulation of the miR-183/96/182 Cluster and the FoxO Family Protein Members Are Not Required for Efficient Replication of HSV-1.Viruses. 2022 Jul 28;14(8):1661. doi: 10.3390/v14081661. Viruses. 2022. PMID: 36016282 Free PMC article.
-
Liver X Receptor-Inducible Host E3 Ligase IDOL Targets a Human Cytomegalovirus Reactivation Determinant.J Virol. 2023 Jul 27;97(7):e0075823. doi: 10.1128/jvi.00758-23. Epub 2023 Jun 20. J Virol. 2023. PMID: 37338407 Free PMC article.
-
Control of Immediate Early Gene Expression for Human Cytomegalovirus Reactivation.Front Cell Infect Microbiol. 2020 Sep 17;10:476. doi: 10.3389/fcimb.2020.00476. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33072616 Free PMC article. Review.
-
NFκB and Cyclic AMP Response Element Sites Mediate the Valproic Acid and UL138 Responsiveness of the Human Cytomegalovirus Major Immediate Early Enhancer and Promoter.J Virol. 2023 Mar 30;97(3):e0002923. doi: 10.1128/jvi.00029-23. Epub 2023 Mar 1. J Virol. 2023. PMID: 36856444 Free PMC article.
References
-
- Nogalski M. T., Collins-McMillen D., Yurochko A. D., Overview of human cytomegalovirus pathogenesis. Methods Mol. Biol. 1119, 15–28 (2014). - PubMed
-
- Sinclair J. H., Baillie J., Bryant L. A., Taylor-Wiedeman J. A., Sissons J. G., Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J. Gen. Virol. 73, 433–435 (1992). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

