Comprehensive mapping of immune perturbations associated with severe COVID-19
- PMID: 32669287
- PMCID: PMC7402634
- DOI: 10.1126/sciimmunol.abd7114
Comprehensive mapping of immune perturbations associated with severe COVID-19
Abstract
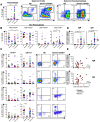
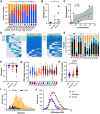
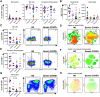
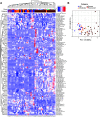
Although critical illness has been associated with SARS-CoV-2-induced hyperinflammation, the immune correlates of severe COVID-19 remain unclear. Here, we comprehensively analyzed peripheral blood immune perturbations in 42 SARS-CoV-2 infected and recovered individuals. We identified extensive induction and activation of multiple immune lineages, including T cell activation, oligoclonal plasmablast expansion, and Fc and trafficking receptor modulation on innate lymphocytes and granulocytes, that distinguished severe COVID-19 cases from healthy donors or SARS-CoV-2-recovered or moderate severity patients. We found the neutrophil to lymphocyte ratio to be a prognostic biomarker of disease severity and organ failure. Our findings demonstrate broad innate and adaptive leukocyte perturbations that distinguish dysregulated host responses in severe SARS-CoV-2 infection and warrant therapeutic investigation.
Copyright © 2020, American Association for the Advancement of Science.
Figures


Similar articles
-
SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function.Nat Immunol. 2020 Nov;21(11):1327-1335. doi: 10.1038/s41590-020-0778-2. Epub 2020 Aug 24. Nat Immunol. 2020. PMID: 32839612 Free PMC article.
-
Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications.Science. 2020 Sep 4;369(6508):eabc8511. doi: 10.1126/science.abc8511. Epub 2020 Jul 15. Science. 2020. PMID: 32669297 Free PMC article.
-
Reappearance of effector T cells is associated with recovery from COVID-19.EBioMedicine. 2020 Jul;57:102885. doi: 10.1016/j.ebiom.2020.102885. Epub 2020 Jul 7. EBioMedicine. 2020. PMID: 32650275 Free PMC article.
-
Covid-19: Perspectives on Innate Immune Evasion.Front Immunol. 2020 Sep 30;11:580641. doi: 10.3389/fimmu.2020.580641. eCollection 2020. Front Immunol. 2020. PMID: 33101306 Free PMC article. Review.
-
Pre-existing immunity to SARS-CoV-2: the knowns and unknowns.Nat Rev Immunol. 2020 Aug;20(8):457-458. doi: 10.1038/s41577-020-0389-z. Nat Rev Immunol. 2020. PMID: 32636479 Free PMC article. Review.
Cited by
-
Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19.Cell. 2021 Apr 1;184(7):1836-1857.e22. doi: 10.1016/j.cell.2021.02.018. Epub 2021 Feb 10. Cell. 2021. PMID: 33713619 Free PMC article.
-
Self-supervised contrastive learning for integrative single cell RNA-seq data analysis.Brief Bioinform. 2022 Sep 20;23(5):bbac377. doi: 10.1093/bib/bbac377. Brief Bioinform. 2022. PMID: 36089561 Free PMC article.
-
Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection.J Exp Med. 2021 May 3;218(5):e20202617. doi: 10.1084/jem.20202617. J Exp Med. 2021. PMID: 33646265 Free PMC article. Clinical Trial.
-
Characterization of T lymphocytes in severe COVID-19 patients.J Med Virol. 2021 Sep;93(9):5608-5613. doi: 10.1002/jmv.27037. Epub 2021 May 3. J Med Virol. 2021. PMID: 33913544 Free PMC article.
-
Heterogenous CD8+ T Cell Maturation and 'Polarization' in Acute and Convalescent COVID-19 Patients.Viruses. 2022 Aug 28;14(9):1906. doi: 10.3390/v14091906. Viruses. 2022. PMID: 36146713 Free PMC article.
References
-
- W. Novel-Coronavirus-2019 Reports. (World Health Organization, 2020), vol. 2020.
-
- Guan W. J., Ni Z. Y., Hu Y., Liang W. H., Ou C. Q., He J. X., Liu L., Shan H., Lei C. L., Hui D. S. C., Du B., Li L. J., Zeng G., Yuen K. Y., Chen R. C., Tang C. L., Wang T., Chen P. Y., Xiang J., Li S. Y., Wang J. L., Liang Z. J., Peng Y. X., Wei L., Liu Y., Hu Y. H., Peng P., Wang J. M., Liu J. Y., Chen Z., Li G., Zheng Z. J., Qiu S. Q., Luo J., Ye C. J., Zhu S. Y., Zhong N. S.; China Medical Treatment Expert Group for Covid-19 , Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). 10.1056/NEJMoa2002032 - DOI - PMC - PubMed
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). 10.1016/S0140-6736(20)30183-5 - DOI - PMC - PubMed
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L., Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 395, 507–513 (2020). 10.1016/S0140-6736(20)30211-7 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19 AI117950/AI/NIAID NIH HHS/United States
- T32 CA009140/CA/NCI NIH HHS/United States
- UM1 AI144288/AI/NIAID NIH HHS/United States
- R01 AI118694/AI/NIAID NIH HHS/United States
- R01 AI115712/AI/NIAID NIH HHS/United States
- P30 CA016520/CA/NCI NIH HHS/United States
- AI117950/NH/NIH HHS/United States
- AI108545/NH/NIH HHS/United States
- P01 CA210944/CA/NCI NIH HHS/United States
- T32 AR076951/AR/NIAMS NIH HHS/United States
- K08 AI136660/AI/NIAID NIH HHS/United States
- AI05343/NH/NIH HHS/United States
- AI115712/NH/NIH HHS/United States
- CA210944/NH/NIH HHS/United States
- AI082630/NH/NIH HHS/United States
- R01 HL137006/HL/NHLBI NIH HHS/United States
- R01 HL137915/HL/NHLBI NIH HHS/United States
- HL137915/NH/NIH HHS/United States
- U19 AI082630/AI/NIAID NIH HHS/United States
- T32 AR007442/AR/NIAMS NIH HHS/United States
- UC4 DK112217/DK/NIDDK NIH HHS/United States
- P01 AI108545/AI/NIAID NIH HHS/United States
- HL137006/NH/NIH HHS/United States
- R01 AI105343/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

