Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions
- PMID: 32653452
- PMCID: PMC7347485
- DOI: 10.1016/j.antiviral.2020.104873
Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions
Abstract
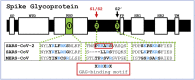
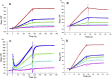
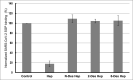
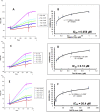
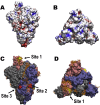
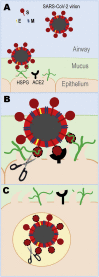
Severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) has resulted in a pandemic and continues to spread around the globe at an unprecedented rate. To date, no effective therapeutic is available to fight its associated disease, COVID-19. Our discovery of a novel insertion of glycosaminoglycan (GAG)-binding motif at S1/S2 proteolytic cleavage site (681-686 (PRRARS)) and two other GAG-binding-like motifs within SARS-CoV-2 spike glycoprotein (SGP) led us to hypothesize that host cell surface GAGs may interact SARS-CoV-2 SGPs to facilitate host cell entry. Using a surface plasmon resonance direct binding assay, we found that both monomeric and trimeric SARS-CoV-2 SGP bind more tightly to immobilized heparin (KD = 40 pM and 73 pM, respectively) than the SARS-CoV and MERS-CoV SGPs (500 nM and 1 nM, respectively). In competitive binding studies, the IC50 of heparin, tri-sulfated non-anticoagulant heparan sulfate, and non-anticoagulant low molecular weight heparin against SARS-CoV-2 SGP binding to immobilized heparin were 0.056 μM, 0.12 μM, and 26.4 μM, respectively. Finally, unbiased computational ligand docking indicates that heparan sulfate interacts with the GAG-binding motif at the S1/S2 site on each monomer interface in the trimeric SARS-CoV-2 SGP, and at another site (453-459 (YRLFRKS)) when the receptor-binding domain is in an open conformation. The current study serves a foundation to further investigate biological roles of GAGs in SARS-CoV-2 pathogenesis. Furthermore, our findings may provide additional basis for further heparin-based interventions for COVID-19 patients exhibiting thrombotic complications.
Keywords: Binding interactions; COVID-19; Glycosaminoglycans; Heparin; SARS-CoV-2; Spike glycoprotein.
Copyright © 2020 Elsevier B.V. All rights reserved.
Figures
Similar articles
-
The structure-activity relationship of the interactions of SARS-CoV-2 spike glycoproteins with glucuronomannan and sulfated galactofucan from Saccharina japonica.Int J Biol Macromol. 2020 Nov 15;163:1649-1658. doi: 10.1016/j.ijbiomac.2020.09.184. Epub 2020 Sep 24. Int J Biol Macromol. 2020. PMID: 32979436 Free PMC article.
-
SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2.Cell. 2020 Nov 12;183(4):1043-1057.e15. doi: 10.1016/j.cell.2020.09.033. Epub 2020 Sep 14. Cell. 2020. PMID: 32970989 Free PMC article.
-
Effective Inhibition of SARS-CoV-2 Entry by Heparin and Enoxaparin Derivatives.J Virol. 2021 Jan 13;95(3):e01987-20. doi: 10.1128/JVI.01987-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33173010 Free PMC article.
-
Molecular Basis of Pathogenesis of Coronaviruses: A Comparative Genomics Approach to Planetary Health to Prevent Zoonotic Outbreaks in the 21st Century.OMICS. 2020 Nov;24(11):634-644. doi: 10.1089/omi.2020.0131. Epub 2020 Sep 16. OMICS. 2020. PMID: 32940573 Review.
-
Angiotensin-converting enzyme 2: The old door for new severe acute respiratory syndrome coronavirus 2 infection.Rev Med Virol. 2020 Sep;30(5):e2122. doi: 10.1002/rmv.2122. Epub 2020 Jun 30. Rev Med Virol. 2020. PMID: 32602627 Free PMC article. Review.
Cited by
-
Impact of Comorbidities on SARS-CoV-2 Viral Entry-Related Genes.J Pers Med. 2020 Sep 25;10(4):146. doi: 10.3390/jpm10040146. J Pers Med. 2020. PMID: 32992731 Free PMC article.
-
Tailor-Made Polysaccharides for Biomedical Applications.ACS Appl Bio Mater. 2024 Jul 15;7(7):4193-4230. doi: 10.1021/acsabm.3c01199. Epub 2024 Jul 3. ACS Appl Bio Mater. 2024. PMID: 38958361 Free PMC article. Review.
-
In silico analysis of dietary polyphenols and their gut microbial metabolites suggest inhibition of SARS-CoV-2 infection, replication, and host inflammatory mediators.J Biomol Struct Dyn. 2023;41(23):14339-14357. doi: 10.1080/07391102.2023.2180669. Epub 2023 Feb 20. J Biomol Struct Dyn. 2023. PMID: 36803516 Free PMC article.
-
The diverse role of heparan sulfate and other GAGs in SARS-CoV-2 infections and therapeutics.Carbohydr Polym. 2023 Jan 1;299:120167. doi: 10.1016/j.carbpol.2022.120167. Epub 2022 Sep 28. Carbohydr Polym. 2023. PMID: 36876764 Free PMC article. Review.
-
Docking and Molecular Dynamics Simulations Clarify Binding Sites for Interactions of Novel Marine Sulfated Glycans with SARS-CoV-2 Spike Glycoprotein.Molecules. 2023 Sep 3;28(17):6413. doi: 10.3390/molecules28176413. Molecules. 2023. PMID: 37687244 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

