Kinetics of viral load and antibody response in relation to COVID-19 severity
- PMID: 32634129
- PMCID: PMC7524490
- DOI: 10.1172/JCI138759
Kinetics of viral load and antibody response in relation to COVID-19 severity
Abstract
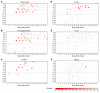
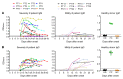
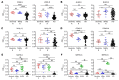
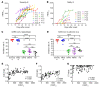
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent for coronavirus 2019 (COVID-19) pneumonia. Little is known about the kinetics, tissue distribution, cross-reactivity, and neutralization antibody response in patients with COVID-19. Two groups of patients with RT-PCR-confirmed COVID-19 were enrolled in this study: 12 severely ill patients in intensive care units who needed mechanical ventilation and 11 mildly ill patients in isolation wards. Serial clinical samples were collected for laboratory detection. Results showed that most of the severely ill patients had viral shedding in a variety of tissues for 20-40 days after onset of disease (8/12, 66.7%), while the majority of mildly ill patients had viral shedding restricted to the respiratory tract and had no detectable virus RNA 10 days after onset (9/11, 81.8%). Mildly ill patients showed significantly lower IgM response compared with that of the severe group. IgG responses were detected in most patients in both the severe and mild groups at 9 days after onset, and remained at a high level throughout the study. Antibodies cross-reactive to SARS-CoV and SARS-CoV-2 were detected in patients with COVID-19 but not in patients with MERS. High levels of neutralizing antibodies were induced after about 10 days after onset in both severely and mildly ill patients which were higher in the severe group. SARS-CoV-2 pseudotype neutralization test and focus reduction neutralization test with authentic virus showed consistent results. Sera from patients with COVID-19 inhibited SARS-CoV-2 entry. Sera from convalescent patients with SARS or Middle East respiratory syndrome (MERS) did not. Anti-SARS-CoV-2 S and N IgG levels exhibited a moderate correlation with neutralization titers in patients' plasma. This study improves our understanding of immune response in humans after SARS-CoV-2 infection.
Keywords: Adaptive immunity; Infectious disease; Molecular biology; Molecular diagnosis; Virology.
Conflict of interest statement
Figures


Comment in
-
SARS-CoV-2 viral load and antibody responses: the case for convalescent plasma therapy.J Clin Invest. 2020 Oct 1;130(10):5112-5114. doi: 10.1172/JCI139760. J Clin Invest. 2020. PMID: 32634126 Free PMC article.
Similar articles
-
SARS-CoV-2 Antibody Responses Do Not Predict COVID-19 Disease Severity.Am J Clin Pathol. 2020 Sep 8;154(4):459-465. doi: 10.1093/ajcp/aqaa123. Am J Clin Pathol. 2020. PMID: 32666092 Free PMC article.
-
Daily Viral Kinetics and Innate and Adaptive Immune Response Assessment in COVID-19: a Case Series.mSphere. 2020 Nov 11;5(6):e00827-20. doi: 10.1128/mSphere.00827-20. mSphere. 2020. PMID: 33177214 Free PMC article.
-
Evaluation of performance of two SARS-CoV-2 Rapid IgM-IgG combined antibody tests on capillary whole blood samples from the fingertip.PLoS One. 2020 Sep 17;15(9):e0237694. doi: 10.1371/journal.pone.0237694. eCollection 2020. PLoS One. 2020. PMID: 32941461 Free PMC article.
-
[SARS-CoV-2 and Microbiological Diagnostic Dynamics in COVID-19 Pandemic].Mikrobiyol Bul. 2020 Jul;54(3):497-509. doi: 10.5578/mb.69839. Mikrobiyol Bul. 2020. PMID: 32755524 Review. Turkish.
-
Differentials of SARS-CoV-2 Viral RNA Re-positivity in Discharged COVID-19 Patients.AIDS Rev. 2021 Jun 3;23(3):153-163. doi: 10.24875/AIDSRev.21000023. AIDS Rev. 2021. PMID: 34082440 Review.
Cited by
-
Severe COVID-19 in HIV/Leishmania infantum coinfected patient: a successfully managed case report.BMC Infect Dis. 2024 Aug 23;24(1):854. doi: 10.1186/s12879-024-09691-5. BMC Infect Dis. 2024. PMID: 39174900 Free PMC article.
-
Titers of IgG and IgA against SARS-CoV-2 proteins and their association with symptoms in mild COVID-19 infection.Sci Rep. 2024 Jun 3;14(1):12725. doi: 10.1038/s41598-024-59634-y. Sci Rep. 2024. PMID: 38830902 Free PMC article.
-
Factors Associated with All-Cause 30-Day Mortality in Indonesian Inpatient COVID-19 Patients at Cipto Mangunkusumo National General Hospital.J Clin Med. 2024 May 20;13(10):2998. doi: 10.3390/jcm13102998. J Clin Med. 2024. PMID: 38792539 Free PMC article.
-
The T-cell response to SARS-CoV-2: kinetic and quantitative aspects and the case for their protective role.Oxf Open Immunol. 2021 Feb 23;2(1):iqab006. doi: 10.1093/oxfimm/iqab006. eCollection 2021. Oxf Open Immunol. 2021. PMID: 38626271 Free PMC article. Review.
-
Comparison of Neutralizing Activity between Vaccinated and Unvaccinated Hospitalized COVID-19 Patients Infected with Delta, Omicron BA.1, or Omicron BA.2 Variant.Microorganisms. 2024 Mar 2;12(3):509. doi: 10.3390/microorganisms12030509. Microorganisms. 2024. PMID: 38543560 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

