The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation
- PMID: 32630219
- PMCID: PMC7411667
- DOI: 10.3390/v12070714
The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation
Abstract
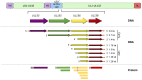
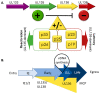
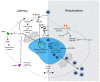
Human cytomegalovirus (HCMV) latency, the means by which the virus persists indefinitely in an infected individual, is a major frontier of current research efforts in the field. Towards developing a comprehensive understanding of HCMV latency and its reactivation from latency, viral determinants of latency and reactivation and their host interactions that govern the latent state and reactivation from latency have been identified. The polycistronic UL133-UL138 locus encodes determinants of both latency and reactivation. In this review, we survey the model systems used to investigate latency and new findings from these systems. Particular focus is given to the roles of the UL133, UL135, UL136 and UL138 proteins in regulating viral latency and how their known host interactions contribute to regulating host signaling pathways towards the establishment of or exit from latency. Understanding the mechanisms underlying viral latency and reactivation is important in developing strategies to block reactivation and prevent CMV disease in immunocompromised individuals, such as transplant patients.
Keywords: DNA virus; Human cytomegalovirus; UL135; UL136; UL138; herpesvirus; latency; reactivation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Stabilization of the human cytomegalovirus UL136p33 reactivation determinant overcomes the requirement for UL135 for replication in hematopoietic cells.J Virol. 2023 Aug 31;97(8):e0014823. doi: 10.1128/jvi.00148-23. Epub 2023 Aug 11. J Virol. 2023. PMID: 37565749 Free PMC article.
-
Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection.J Virol. 2014 Jun;88(11):5987-6002. doi: 10.1128/JVI.03506-13. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623432 Free PMC article.
-
Complex Interplay of the UL136 Isoforms Balances Cytomegalovirus Replication and Latency.mBio. 2016 Mar 1;7(2):e01986. doi: 10.1128/mBio.01986-15. mBio. 2016. PMID: 26933055 Free PMC article.
-
Human cytomegalovirus manipulation of latently infected cells.Viruses. 2013 Nov 21;5(11):2803-24. doi: 10.3390/v5112803. Viruses. 2013. PMID: 24284875 Free PMC article. Review.
-
[Research Progress in Mechanisms Associated with Latent Infection in Humans by the Cytomegalovirus].Bing Du Xue Bao. 2016 Jan;32(1):82-7. Bing Du Xue Bao. 2016. PMID: 27295888 Review. Chinese.
Cited by
-
Autophagy in Viral Development and Progression of Cancer.Front Oncol. 2021 Mar 8;11:603224. doi: 10.3389/fonc.2021.603224. eCollection 2021. Front Oncol. 2021. PMID: 33763351 Free PMC article. Review.
-
The Human Cytomegalovirus Latency-Associated Gene Product Latency Unique Natural Antigen Regulates Latent Gene Expression.Viruses. 2023 Sep 4;15(9):1875. doi: 10.3390/v15091875. Viruses. 2023. PMID: 37766281 Free PMC article.
-
Modulation of host cell signaling during cytomegalovirus latency and reactivation.Virol J. 2021 Oct 18;18(1):207. doi: 10.1186/s12985-021-01674-1. Virol J. 2021. PMID: 34663377 Free PMC article. Review.
-
HCMV Antivirals and Strategies to Target the Latent Reservoir.Viruses. 2021 May 1;13(5):817. doi: 10.3390/v13050817. Viruses. 2021. PMID: 34062863 Free PMC article. Review.
-
Sirtuin 2 promotes human cytomegalovirus replication by regulating cell cycle progression.mSystems. 2023 Dec 21;8(6):e0051023. doi: 10.1128/msystems.00510-23. Epub 2023 Nov 2. mSystems. 2023. PMID: 37916830 Free PMC article.
References
-
- Britt W. Manifestations of Human Cytomegalovirus Infection: Proposed Mechanisms of Acute and Chronic Disease. In: Shenk T.E., Stinski M.F., editors. Human Cytomegalovirus. Springer; Berlin/Heidelberg, Germany: 2008. pp. 417–470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

