Cell-to-cell expression dispersion of B-cell surface proteins is linked to genetic variants in humans
- PMID: 32620900
- PMCID: PMC7335051
- DOI: 10.1038/s42003-020-1075-1
Cell-to-cell expression dispersion of B-cell surface proteins is linked to genetic variants in humans
Abstract
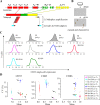
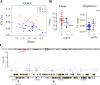
Variability in gene expression across a population of homogeneous cells is known to influence various biological processes. In model organisms, natural genetic variants were found that modify expression dispersion (variability at a fixed mean) but very few studies have detected such effects in humans. Here, we analyzed single-cell expression of four proteins (CD23, CD55, CD63 and CD86) across cell lines derived from individuals of the Yoruba population. Using data from over 30 million cells, we found substantial inter-individual variation of dispersion. We demonstrate, via de novo cell line generation and subcloning experiments, that this variation exceeds the variation associated with cellular immortalization. We detected a genetic association between the expression dispersion of CD63 and the rs971 SNP. Our results show that human DNA variants can have inherently-probabilistic effects on gene expression. Such subtle genetic effects may participate to phenotypic variation and disease outcome.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus.Nat Genet. 2008 Feb;40(2):211-6. doi: 10.1038/ng.79. Epub 2008 Jan 20. Nat Genet. 2008. PMID: 18204447
-
Detection of high variability in gene expression from single-cell RNA-seq profiling.BMC Genomics. 2016 Aug 22;17 Suppl 7(Suppl 7):508. doi: 10.1186/s12864-016-2897-6. BMC Genomics. 2016. PMID: 27556924 Free PMC article.
-
The cis and trans effects of the risk variants of coronary artery disease in the Chr9p21 region.BMC Med Genomics. 2015 May 10;8:21. doi: 10.1186/s12920-015-0094-0. BMC Med Genomics. 2015. PMID: 25958224 Free PMC article.
-
Large-scale profiling and identification of potential regulatory mechanisms for allelic gene expression in colorectal cancer cells.Gene. 2013 Jan 1;512(1):16-22. doi: 10.1016/j.gene.2012.10.001. Epub 2012 Oct 10. Gene. 2013. PMID: 23064046
-
Endometrial vezatin and its association with endometriosis risk.Hum Reprod. 2016 May;31(5):999-1013. doi: 10.1093/humrep/dew047. Epub 2016 Mar 22. Hum Reprod. 2016. PMID: 27005890
References
-
- Oates AC. What’s all the noise about developmental stochasticity? Development. 2011;138:601–607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

