JMJD6 participates in the maintenance of ribosomal DNA integrity in response to DNA damage
- PMID: 32598339
- PMCID: PMC7351224
- DOI: 10.1371/journal.pgen.1008511
JMJD6 participates in the maintenance of ribosomal DNA integrity in response to DNA damage
Abstract
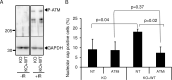
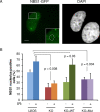
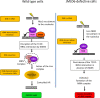
Ribosomal DNA (rDNA) is the most transcribed genomic region and contains hundreds of tandem repeats. Maintaining these rDNA repeats as well as the level of rDNA transcription is essential for cellular homeostasis. DNA damages generated in rDNA need to be efficiently and accurately repaired and rDNA repeats instability has been reported in cancer, aging and neurological diseases. Here, we describe that the histone demethylase JMJD6 is rapidly recruited to nucleolar DNA damage and is crucial for the relocalisation of rDNA in nucleolar caps. Yet, JMJD6 is dispensable for rDNA transcription inhibition. Mass spectrometry analysis revealed that JMJD6 interacts with the nucleolar protein Treacle and modulates its interaction with NBS1. Moreover, cells deficient for JMJD6 show increased sensitivity to nucleolar DNA damage as well as loss and rearrangements of rDNA repeats upon irradiation. Altogether our data reveal that rDNA transcription inhibition is uncoupled from rDNA relocalisation into nucleolar caps and that JMJD6 is required for rDNA stability through its role in nucleolar caps formation.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures
Similar articles
-
Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair.Nucleic Acids Res. 2019 Sep 5;47(15):8019-8035. doi: 10.1093/nar/gkz518. Nucleic Acids Res. 2019. PMID: 31184714 Free PMC article.
-
Treacle controls the nucleolar response to rDNA breaks via TOPBP1 recruitment and ATR activation.Nat Commun. 2020 Jan 8;11(1):123. doi: 10.1038/s41467-019-13981-x. Nat Commun. 2020. PMID: 31913317 Free PMC article.
-
JMJD6 modulates DNA damage response through downregulating H4K16ac independently of its enzymatic activity.Cell Death Differ. 2020 Mar;27(3):1052-1066. doi: 10.1038/s41418-019-0397-3. Epub 2019 Jul 29. Cell Death Differ. 2020. PMID: 31358914 Free PMC article.
-
[A CxxC domain that binds to unmethylated CpG is required for KDM2A to control rDNA transcription].Yakugaku Zasshi. 2015;135(1):11-21. doi: 10.1248/yakushi.14-00202-2. Yakugaku Zasshi. 2015. PMID: 25743893 Review. Japanese.
-
Jumonji domain-containing protein 6 protein and its role in cancer.Cell Prolif. 2020 Feb;53(2):e12747. doi: 10.1111/cpr.12747. Epub 2020 Jan 21. Cell Prolif. 2020. PMID: 31961032 Free PMC article. Review.
Cited by
-
Treacle Sticks the Nucleolar Responses to DNA Damage Together.Front Cell Dev Biol. 2022 May 12;10:892006. doi: 10.3389/fcell.2022.892006. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35646927 Free PMC article. Review.
-
Treacle and TOPBP1 control replication stress response in the nucleolus.J Cell Biol. 2021 Aug 2;220(8):e202008085. doi: 10.1083/jcb.202008085. Epub 2021 Jun 8. J Cell Biol. 2021. PMID: 34100862 Free PMC article.
-
To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families.Int J Mol Sci. 2024 Jun 24;25(13):6900. doi: 10.3390/ijms25136900. Int J Mol Sci. 2024. PMID: 39000010 Free PMC article. Review.
-
Widespread hydroxylation of unstructured lysine-rich protein domains by JMJD6.Proc Natl Acad Sci U S A. 2022 Aug 9;119(32):e2201483119. doi: 10.1073/pnas.2201483119. Epub 2022 Aug 5. Proc Natl Acad Sci U S A. 2022. PMID: 35930668 Free PMC article.
-
53BP1-mediated recruitment of RASSF1A to ribosomal DNA breaks promotes local ATM signaling.EMBO Rep. 2022 Aug 3;23(8):e54483. doi: 10.15252/embr.202154483. Epub 2022 Jun 27. EMBO Rep. 2022. PMID: 35758159 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

