Sedentary and Trained Older Men Have Distinct Circulating Exosomal microRNA Profiles at Baseline and in Response to Acute Exercise
- PMID: 32587527
- PMCID: PMC7298138
- DOI: 10.3389/fphys.2020.00605
Sedentary and Trained Older Men Have Distinct Circulating Exosomal microRNA Profiles at Baseline and in Response to Acute Exercise
Abstract
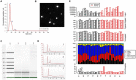
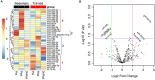
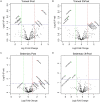
Exercise has multi-systemic benefits and attenuates the physiological impairments associated with aging. Emerging evidence suggests that circulating exosomes mediate some of the beneficial effects of exercise via the transfer of microRNAs between tissues. However, the impact of regular exercise and acute exercise on circulating exosomal microRNAs (exomiRs) in older populations remains unknown. In the present study, we analyzed circulating exomiR expression in endurance-trained elderly men (n = 5) and age-matched sedentary males (n = 5) at baseline (Pre), immediately after a forty minute bout of aerobic exercise on a cycle ergometer (Post), and three hours after this acute exercise (3hPost). Following the isolation and enrichment of exosomes from plasma, exosome-enriched preparations were characterized and exomiR levels were determined by sequencing. The effect of regular exercise on circulating exomiRs was assessed by comparing the baseline expression levels in the trained and sedentary groups. The effect of acute exercise was determined by comparing baseline and post-training expression levels in each group. Regular exercise resulted in significantly increased baseline expression of three exomiRs (miR-486-5p, miR-215-5p, miR-941) and decreased expression of one exomiR (miR-151b). Acute exercise altered circulating exomiR expression in both groups. However, exomiRs regulated by acute exercise in the trained group (7 miRNAs at Post and 8 at 3hPost) were distinct from those in the sedentary group (9 at Post and 4 at 3hPost). Pathway analysis prediction and reported target validation experiments revealed that the majority of exercise-regulated exomiRs are targeting genes that are related to IGF-1 signaling, a pathway involved in exercise-induced muscle and cardiac hypertrophy. The immediately post-acute exercise exomiR signature in the trained group correlates with activation of IGF-1 signaling, whereas in the sedentary group it is associated with inhibition of IGF-1 signaling. While further validation is needed, including measurements of IGF-1/IGF-1 signaling in blood or skeletal muscle, our results suggest that training status may counteract age-related anabolic resistance by modulating circulating exomiR profiles both at baseline and in response to acute exercise.
Keywords: Acute aerobic exercise; aging; insulin growth factor-1 signaling; microRNA profiling; plasma exosomes; regular endurance training.
Copyright © 2020 Nair, Ge, Li, Pincas, Jain, Seenarine, Amper, Goodpaster, Walsh, Coen and Sealfon.
Figures
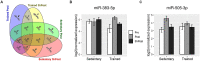
Similar articles
-
Physical Activity as a Preventive Lifestyle Intervention Acts Through Specific Exosomal miRNA Species-Evidence From Human Short- and Long-Term Pilot Studies.Front Physiol. 2021 Aug 2;12:658218. doi: 10.3389/fphys.2021.658218. eCollection 2021. Front Physiol. 2021. PMID: 34408656 Free PMC article.
-
Using bioinformatics technology to mine the expression of serum exosomal miRNA in patients with traumatic brain injury.Front Neurosci. 2023 Apr 18;17:1145307. doi: 10.3389/fnins.2023.1145307. eCollection 2023. Front Neurosci. 2023. PMID: 37144089 Free PMC article.
-
Longterm Exercise-Derived Exosomal miR-342-5p: A Novel Exerkine for Cardioprotection.Circ Res. 2019 Apr 26;124(9):1386-1400. doi: 10.1161/CIRCRESAHA.118.314635. Circ Res. 2019. PMID: 30879399
-
Prospecting of exosomal-miRNA signatures as prognostic marker for gestational diabetes mellitus and other adverse pregnancy outcomes.Front Endocrinol (Lausanne). 2023 Feb 9;14:1097337. doi: 10.3389/fendo.2023.1097337. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36843574 Free PMC article. Review.
-
Exosomal microRNAs (exomiRs): Small molecules with a big role in cancer.Cancer Lett. 2018 Apr 28;420:228-235. doi: 10.1016/j.canlet.2018.02.002. Epub 2018 Feb 7. Cancer Lett. 2018. PMID: 29425686 Free PMC article. Review.
Cited by
-
Exercise mimetics: harnessing the therapeutic effects of physical activity.Nat Rev Drug Discov. 2021 Nov;20(11):862-879. doi: 10.1038/s41573-021-00217-1. Epub 2021 Jun 8. Nat Rev Drug Discov. 2021. PMID: 34103713 Review.
-
Regulation of microRNAs in Satellite Cell Renewal, Muscle Function, Sarcopenia and the Role of Exercise.Int J Mol Sci. 2020 Sep 14;21(18):6732. doi: 10.3390/ijms21186732. Int J Mol Sci. 2020. PMID: 32937893 Free PMC article. Review.
-
Role of MicroRNAs and Long Non-Coding RNAs in Sarcopenia.Cells. 2022 Jan 6;11(2):187. doi: 10.3390/cells11020187. Cells. 2022. PMID: 35053303 Free PMC article. Review.
-
Effects of Voluntary Running Wheel Exercise-Induced Extracellular Vesicles on Anxiety.Front Mol Neurosci. 2021 Jul 1;14:665800. doi: 10.3389/fnmol.2021.665800. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34276303 Free PMC article.
-
Potential involvement of circulating extracellular vesicles and particles on exercise effects in malignancies.Front Endocrinol (Lausanne). 2023 Mar 3;14:1121390. doi: 10.3389/fendo.2023.1121390. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36936170 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

