Talin dissociates from RIAM and associates to vinculin sequentially in response to the actomyosin force
- PMID: 32561773
- PMCID: PMC7305319
- DOI: 10.1038/s41467-020-16922-1
Talin dissociates from RIAM and associates to vinculin sequentially in response to the actomyosin force
Abstract
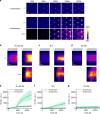
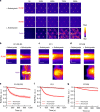
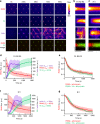
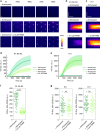
Cells reinforce adhesion strength and cytoskeleton anchoring in response to the actomyosin force. The mechanical stretching of talin, which exposes cryptic vinculin-binding sites, triggers this process. The binding of RIAM to talin could regulate this mechanism. However, the mechanosensitivity of the talin-RIAM complex has never been tested. It is also not known whether RIAM controls the mechanosensitivity of the talin-vinculin complex. To address these issues, we designed an in vitro microscopy assay with purified proteins in which the actomyosin force controls RIAM and vinculin-binding to talin. We demonstrate that actomyosin triggers RIAM dissociation from several talin domains. Actomyosin also provokes the sequential exchange of RIAM for vinculin on talin. The effect of RIAM on this force-dependent binding of vinculin to talin varies from one talin domain to another. This mechanism could allow talin to biochemically code a wide range of forces by selecting different combinations of partners.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Actomyosin-dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring.Nat Commun. 2014;5:3095. doi: 10.1038/ncomms4095. Nat Commun. 2014. PMID: 24452080 Free PMC article.
-
RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover.J Biol Chem. 2013 Mar 22;288(12):8238-8249. doi: 10.1074/jbc.M112.438119. Epub 2013 Feb 6. J Biol Chem. 2013. PMID: 23389036 Free PMC article.
-
Pressure-Dependent Chemical Shifts in the R3 Domain of Talin Show that It Is Thermodynamically Poised for Binding to Either Vinculin or RIAM.Structure. 2017 Dec 5;25(12):1856-1866.e2. doi: 10.1016/j.str.2017.10.008. Epub 2017 Nov 16. Structure. 2017. PMID: 29153504
-
The Rap1-RIAM-talin axis of integrin activation and blood cell function.Blood. 2016 Jul 28;128(4):479-87. doi: 10.1182/blood-2015-12-638700. Epub 2016 May 20. Blood. 2016. PMID: 27207789 Free PMC article. Review.
-
Integrin activation.Biochem Soc Trans. 2008 Apr;36(Pt 2):229-34. doi: 10.1042/BST0360229. Biochem Soc Trans. 2008. PMID: 18363565 Free PMC article. Review.
Cited by
-
LIM domain proteins in cell mechanobiology.Cytoskeleton (Hoboken). 2021 Jun;78(6):303-311. doi: 10.1002/cm.21677. Epub 2021 Jun 10. Cytoskeleton (Hoboken). 2021. PMID: 34028199 Free PMC article. Review.
-
Structural, biochemical, and functional properties of the Rap1-Interacting Adaptor Molecule (RIAM).Biomed J. 2022 Apr;45(2):289-298. doi: 10.1016/j.bj.2021.09.005. Epub 2021 Oct 1. Biomed J. 2022. PMID: 34601137 Free PMC article. Review.
-
Expression of the phagocytic receptors αMβ2 and αXβ2 is controlled by RIAM, VASP and Vinculin in neutrophil-differentiated HL-60 cells.Front Immunol. 2022 Sep 27;13:951280. doi: 10.3389/fimmu.2022.951280. eCollection 2022. Front Immunol. 2022. PMID: 36238292 Free PMC article.
-
Manipulation of Focal Adhesion Signaling by Pathogenic Microbes.Int J Mol Sci. 2021 Jan 29;22(3):1358. doi: 10.3390/ijms22031358. Int J Mol Sci. 2021. PMID: 33572997 Free PMC article. Review.
-
Talin‑1 interaction network in cellular mechanotransduction (Review).Int J Mol Med. 2022 May;49(5):60. doi: 10.3892/ijmm.2022.5116. Epub 2022 Mar 10. Int J Mol Med. 2022. PMID: 35266014 Free PMC article. Review.
References
-
- Le Clainche C, Carlier M-F. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 2008;88:489–513. - PubMed
-
- Wehrle-Haller B. Structure and function of focal adhesions. Curr. Opin. Cell Biol. 2012;24:116–124. - PubMed
-
- Ciobanasu C, Faivre B, Le Clainche C. Integrating actin dynamics, mechanotransduction and integrin activation: the multiple functions of actin binding proteins in focal adhesions. Eur. J. Cell Biol. 2013;92:339–348. - PubMed
-
- Bachmann M, Kukkurainen S, Hytönen VP, Wehrle-Haller B. Cell adhesion by integrins. Physiol. Rev. 2019;99:1655–1699. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

