Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals
- PMID: 32473127
- PMCID: PMC7237901
- DOI: 10.1016/j.cell.2020.05.015
Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals
Abstract
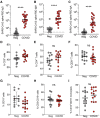
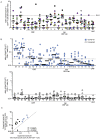
Understanding adaptive immunity to SARS-CoV-2 is important for vaccine development, interpreting coronavirus disease 2019 (COVID-19) pathogenesis, and calibration of pandemic control measures. Using HLA class I and II predicted peptide "megapools," circulating SARS-CoV-2-specific CD8+ and CD4+ T cells were identified in ∼70% and 100% of COVID-19 convalescent patients, respectively. CD4+ T cell responses to spike, the main target of most vaccine efforts, were robust and correlated with the magnitude of the anti-SARS-CoV-2 IgG and IgA titers. The M, spike, and N proteins each accounted for 11%-27% of the total CD4+ response, with additional responses commonly targeting nsp3, nsp4, ORF3a, and ORF8, among others. For CD8+ T cells, spike and M were recognized, with at least eight SARS-CoV-2 ORFs targeted. Importantly, we detected SARS-CoV-2-reactive CD4+ T cells in ∼40%-60% of unexposed individuals, suggesting cross-reactive T cell recognition between circulating "common cold" coronaviruses and SARS-CoV-2.
Keywords: CD4; CD8; COVID-19; SARS-CoV-2; T cells; coronavirus; cross-reactivity; epitopes.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures











Comment in
-
Mapping the T cell response to COVID-19.Signal Transduct Target Ther. 2020 Jul 2;5(1):112. doi: 10.1038/s41392-020-00228-1. Signal Transduct Target Ther. 2020. PMID: 32616709 Free PMC article. No abstract available.
-
Characteristics of SARS-CoV-2-specific cytotoxic T cells revealed by single-cell immune profiling of longitudinal COVID-19 blood samples.Signal Transduct Target Ther. 2020 Dec 4;5(1):285. doi: 10.1038/s41392-020-00425-y. Signal Transduct Target Ther. 2020. PMID: 33277469 Free PMC article. No abstract available.
Similar articles
-
Suboptimal SARS-CoV-2-specific CD8+ T cell response associated with the prominent HLA-A*02:01 phenotype.Proc Natl Acad Sci U S A. 2020 Sep 29;117(39):24384-24391. doi: 10.1073/pnas.2015486117. Epub 2020 Sep 10. Proc Natl Acad Sci U S A. 2020. PMID: 32913053 Free PMC article.
-
Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19.Nat Immunol. 2020 Nov;21(11):1336-1345. doi: 10.1038/s41590-020-0782-6. Epub 2020 Sep 4. Nat Immunol. 2020. PMID: 32887977 Free PMC article.
-
Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein.Immunity. 2020 Nov 17;53(5):1095-1107.e3. doi: 10.1016/j.immuni.2020.10.006. Epub 2020 Oct 20. Immunity. 2020. PMID: 33128877 Free PMC article.
-
Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country.Front Immunol. 2020 Sep 3;11:2008. doi: 10.3389/fimmu.2020.02008. eCollection 2020. Front Immunol. 2020. PMID: 33013857 Free PMC article. Review.
-
Cross-reactive memory T cells and herd immunity to SARS-CoV-2.Nat Rev Immunol. 2020 Nov;20(11):709-713. doi: 10.1038/s41577-020-00460-4. Epub 2020 Oct 6. Nat Rev Immunol. 2020. PMID: 33024281 Free PMC article. Review.
Cited by
-
High protection and transmission-blocking immunity elicited by single-cycle SARS-CoV-2 vaccine in hamsters.NPJ Vaccines. 2024 Oct 30;9(1):206. doi: 10.1038/s41541-024-00992-z. NPJ Vaccines. 2024. PMID: 39472701 Free PMC article.
-
T-Cell Immune Responses to SARS-CoV-2 Infection and Vaccination.Vaccines (Basel). 2024 Sep 30;12(10):1126. doi: 10.3390/vaccines12101126. Vaccines (Basel). 2024. PMID: 39460293 Free PMC article. Review.
-
Activation-Induced Marker Assay to Identify and Isolate HCV-Specific T Cells for Single-Cell RNA-Seq Analysis.Viruses. 2024 Oct 17;16(10):1623. doi: 10.3390/v16101623. Viruses. 2024. PMID: 39459954 Free PMC article.
-
T Cell Peptide Prediction, Immune Response, and Host-Pathogen Relationship in Vaccinated and Recovered from Mild COVID-19 Subjects.Biomolecules. 2024 Sep 26;14(10):1217. doi: 10.3390/biom14101217. Biomolecules. 2024. PMID: 39456150 Free PMC article.
-
Investigation of Long-Term CD4+ T Cell Receptor Repertoire Changes Following SARS-CoV-2 Infection in Patients with Different Severities of Disease.Diagnostics (Basel). 2024 Oct 19;14(20):2330. doi: 10.3390/diagnostics14202330. Diagnostics (Basel). 2024. PMID: 39451653 Free PMC article.
References
-
- Bancroft T., Dillon M.B., da Silva Antunes R., Paul S., Peters B., Crotty S., Lindestam Arlehamn C.S., Sette A. Th1 versus Th2 T cell polarization by whole-cell and acellular childhood pertussis vaccines persists upon re-immunization in adolescence and adulthood. Cell. Immunol. 2016;304-305:35–43. - PMC - PubMed
-
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R.A., tenOever B.R. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv. 2020 doi: 10.1101/2020.03.24.004655. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

